Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
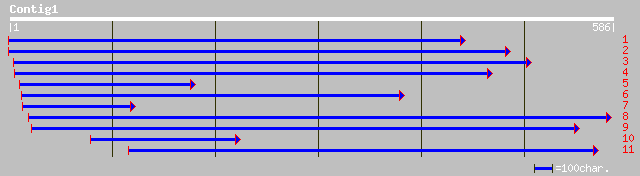
Query= KMC002937A_C01 KMC002937A_c01
(583 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_177517.1| putative RING zinc finger protein; protein id: ... 139 3e-32
pir||A86315 F2H15.19 protein - Arabidopsis thaliana gi|9665074|g... 134 8e-31
gb|AAM63662.1| zinc-finger protein (C-terminal), putative [Arabi... 134 8e-31
ref|NP_173239.1| zinc-finger protein (C-terminal), putative; pro... 134 8e-31
ref|NP_195130.1| putative protein; protein id: At4g34040.1 [Arab... 121 7e-27
>ref|NP_177517.1| putative RING zinc finger protein; protein id: At1g73760.1
[Arabidopsis thaliana] gi|25406292|pir||H96764 protein
RING zinc finger protein F25P22.18 [imported] -
Arabidopsis thaliana
gi|12324210|gb|AAG52076.1|AC012679_14 putative RING zinc
finger protein; 69105-67310 [Arabidopsis thaliana]
Length = 367
Score = 139 bits (350), Expect = 3e-32
Identities = 65/110 (59%), Positives = 84/110 (76%)
Frame = -3
Query: 581 MGGRLNSHDQFRDWRLDIDNMSYEQLLELGEKIGYVNTGVKEDEMELNIRKLKLFISNDT 402
MGG L+S DQFRD RL++DNM+YEQLLELGE+IG+VNTG+ E +++ +RK+K + T
Sbjct: 254 MGGVLSSFDQFRDMRLNVDNMTYEQLLELGERIGHVNTGLTEKQIKSCLRKVKPCRQDTT 313
Query: 401 SKHQLDRKCTICQEEYESDDELGKLKCEHCFHFQCIKQWLVLKNFCPVCK 252
DRKC ICQ+EYE+ DE+G+L+C H FH C+ QWLV KN CPVCK
Sbjct: 314 VA---DRKCIICQDEYEAKDEVGELRCGHRFHIDCVNQWLVRKNSCPVCK 360
>pir||A86315 F2H15.19 protein - Arabidopsis thaliana
gi|9665074|gb|AAF97276.1|AC034106_19 Contains similarity
to RING-H2 finger protein RHG1a (partial) from
Arabidopsis thaliana gb|AF079183 and contains a Zinc
finger (C3HC4 type) PF|00097 domain. EST gb|AV522036
comes from this gene
Length = 383
Score = 134 bits (337), Expect = 8e-31
Identities = 61/111 (54%), Positives = 82/111 (72%)
Frame = -3
Query: 581 MGGRLNSHDQFRDWRLDIDNMSYEQLLELGEKIGYVNTGVKEDEMELNIRKLKLFISNDT 402
MG +S D + + RLD+D+MSYEQLLELG++IGYVNTG+KE E+ + K+K +S+
Sbjct: 271 MGRITDSRDNYHELRLDVDSMSYEQLLELGDRIGYVNTGLKESEIHRCLGKIKPSVSHTL 330
Query: 401 SKHQLDRKCTICQEEYESDDELGKLKCEHCFHFQCIKQWLVLKNFCPVCKQ 249
+DRKC+ICQ+EYE +DE+G+L C H FH C+KQWL KN CPVCK+
Sbjct: 331 ----VDRKCSICQDEYEREDEVGELNCGHSFHVHCVKQWLSRKNACPVCKK 377
>gb|AAM63662.1| zinc-finger protein (C-terminal), putative [Arabidopsis thaliana]
Length = 368
Score = 134 bits (337), Expect = 8e-31
Identities = 61/111 (54%), Positives = 82/111 (72%)
Frame = -3
Query: 581 MGGRLNSHDQFRDWRLDIDNMSYEQLLELGEKIGYVNTGVKEDEMELNIRKLKLFISNDT 402
MG +S D + + RLD+D+MSYEQLLELG++IGYVNTG+KE E+ + K+K +S+
Sbjct: 256 MGRITDSRDNYHELRLDVDSMSYEQLLELGDRIGYVNTGLKESEIHRCLGKIKPSVSHTL 315
Query: 401 SKHQLDRKCTICQEEYESDDELGKLKCEHCFHFQCIKQWLVLKNFCPVCKQ 249
+DRKC+ICQ+EYE +DE+G+L C H FH C+KQWL KN CPVCK+
Sbjct: 316 ----VDRKCSICQDEYEREDEVGELNCGHSFHVHCVKQWLSRKNACPVCKK 362
>ref|NP_173239.1| zinc-finger protein (C-terminal), putative; protein id:
At1g17970.1, supported by cDNA: 261272., supported by
cDNA: gi_16226278 [Arabidopsis thaliana]
gi|16226279|gb|AAL16122.1|AF428290_1 At1g17970/F2H15_16
[Arabidopsis thaliana] gi|21436039|gb|AAM51597.1|
At1g17970/F2H15_16 [Arabidopsis thaliana]
Length = 368
Score = 134 bits (337), Expect = 8e-31
Identities = 61/111 (54%), Positives = 82/111 (72%)
Frame = -3
Query: 581 MGGRLNSHDQFRDWRLDIDNMSYEQLLELGEKIGYVNTGVKEDEMELNIRKLKLFISNDT 402
MG +S D + + RLD+D+MSYEQLLELG++IGYVNTG+KE E+ + K+K +S+
Sbjct: 256 MGRITDSRDNYHELRLDVDSMSYEQLLELGDRIGYVNTGLKESEIHRCLGKIKPSVSHTL 315
Query: 401 SKHQLDRKCTICQEEYESDDELGKLKCEHCFHFQCIKQWLVLKNFCPVCKQ 249
+DRKC+ICQ+EYE +DE+G+L C H FH C+KQWL KN CPVCK+
Sbjct: 316 ----VDRKCSICQDEYEREDEVGELNCGHSFHVHCVKQWLSRKNACPVCKK 362
>ref|NP_195130.1| putative protein; protein id: At4g34040.1 [Arabidopsis thaliana]
gi|7486285|pir||T05432 hypothetical protein F28A23.200 -
Arabidopsis thaliana gi|2911058|emb|CAA17568.1| putative
protein [Arabidopsis thaliana]
gi|7270353|emb|CAB80121.1| putative protein [Arabidopsis
thaliana]
Length = 666
Score = 121 bits (303), Expect = 7e-27
Identities = 57/108 (52%), Positives = 71/108 (64%)
Frame = -3
Query: 575 GRLNSHDQFRDWRLDIDNMSYEQLLELGEKIGYVNTGVKEDEMELNIRKLKLFISNDTSK 396
G HD+ RD RLD+DNMSYE+LL LGE+IG V+TG+ E E+ L + K S+
Sbjct: 554 GMAEMHDRHRDMRLDVDNMSYEELLALGERIGDVSTGLSE-EVILKVMKQHKHTSSAAGS 612
Query: 395 HQLDRKCTICQEEYESDDELGKLKCEHCFHFQCIKQWLVLKNFCPVCK 252
HQ C +CQEEY D+LG L C H FH C+KQWL+LKN CP+CK
Sbjct: 613 HQDMEPCCVCQEEYAEGDDLGTLGCGHEFHTACVKQWLMLKNLCPICK 660
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,841,002
Number of Sequences: 1393205
Number of extensions: 9415965
Number of successful extensions: 31489
Number of sequences better than 10.0: 1182
Number of HSP's better than 10.0 without gapping: 29674
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31272
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)