Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002922A_C09 KMC002922A_c09
(1162 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
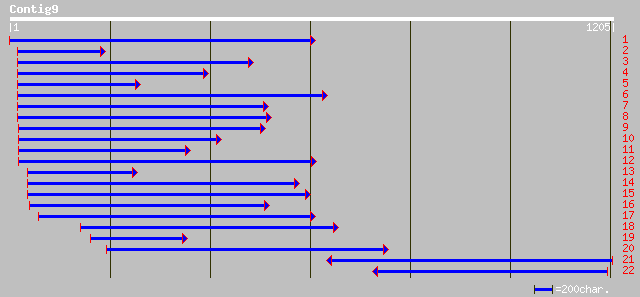
sp|Q01197|E6_GOSHI Protein E6 gi|421806|pir||A46130 fiber protei... 141 2e-32
gb|AAA33056.1| 5' start site is putative; putative 141 2e-32
pir||S65063 fiber protein E6 (clones SIE6-2A and SIE6-3B) - sea-... 136 7e-31
pir||S65062 fiber protein E6 (clone CKE6-4A) - upland cotton gi|... 126 6e-28
pir||T10265 arabinogalactan-protein AGP2 - Persian tobacco gi|10... 114 3e-24
>sp|Q01197|E6_GOSHI Protein E6 gi|421806|pir||A46130 fiber protein E6 (clone CKE6-1A) -
upland cotton gi|167323|gb|AAA33055.1| 5' start site is
putative; putative gi|1000084|gb|AAB03079.1| E6
Length = 238
Score = 141 bits (356), Expect = 2e-32
Identities = 115/301 (38%), Positives = 147/301 (48%), Gaps = 7/301 (2%)
Frame = -1
Query: 1147 LKILFFLLLNNPVDCPMQINARDSQFFSKVTHVNNNNNNVKETEFPNNNEVP--VNKPEQ 974
+ ILF L + MQI+AR+ +FSK VN N E + VP KPE+
Sbjct: 10 MSILFLFALFS-----MQIHARE--YFSKFPRVNINEKETTTREQKHETFVPQTTQKPEE 62
Query: 973 Q-PVFTPETENSYGLYGHESGLHPPTTTT--TYLPYKTTPSKEDHTTTKNFNNNYNNYQK 803
Q P F PET+N YGLYGHESG P+ TT TY PY TP + +
Sbjct: 63 QEPRFIPETQNGYGLYGHESGSSRPSFTTKETYEPY-VTPVR---------------FHP 106
Query: 802 DDFFNTNQNDQLSDTRFIGNSHSSSNNNNYYYNNKNAYEGN-QNELSNTKFTEGGNKYNP 626
D+ +N+ SSNN + YY NKNAYE Q L FTE G ++
Sbjct: 107 DEPYNSIPE--------------SSNNKDTYYYNKNAYESTKQQNLGEAIFTEKG--WST 150
Query: 625 RENQNYNQKFFYNNNAAANENYNFNNNNAANNNEKGAEVQGMSDTRFMEGGKYFYNVNSE 446
+ENQN N +N NN NN EK QGMSDTR++E GKY+Y+V SE
Sbjct: 151 KENQNNNY---------------YNGNNGYNNGEK----QGMSDTRYLENGKYYYDVKSE 191
Query: 445 NEKYNPTGYGGESSTRGVNSENWYNNKGYFGNNNNENKNSMEGY-KNQEEFEDDPDVDFE 269
N Y P + ++RGV S N +N Y N+M Y +NQEEFE+ + +FE
Sbjct: 192 N-NYYPNRF---DNSRGVASRNEFNENRY---------NNMGRYHQNQEEFEESEE-EFE 237
Query: 268 P 266
P
Sbjct: 238 P 238
>gb|AAA33056.1| 5' start site is putative; putative
Length = 218
Score = 141 bits (355), Expect = 2e-32
Identities = 111/285 (38%), Positives = 141/285 (48%), Gaps = 7/285 (2%)
Frame = -1
Query: 1099 MQINARDSQFFSKVTHVNNNNNNVKETEFPNNNEVP--VNKPEQQ-PVFTPETENSYGLY 929
MQI+AR+ +FSK VN N E + VP KPE+Q P F PET+N YGLY
Sbjct: 1 MQIHARE--YFSKFPRVNINEKETTTREQKHETFVPQTTQKPEEQEPRFIPETQNGYGLY 58
Query: 928 GHESGLHPPTTTT--TYLPYKTTPSKEDHTTTKNFNNNYNNYQKDDFFNTNQNDQLSDTR 755
GHESG P+ TT TY PY TP + + D+ +N+
Sbjct: 59 GHESGSSRPSFTTKETYEPY-VTPVR---------------FHPDEPYNSIPE------- 95
Query: 754 FIGNSHSSSNNNNYYYNNKNAYEGN-QNELSNTKFTEGGNKYNPRENQNYNQKFFYNNNA 578
SSNN + YY NKNAYE Q L FTE G ++ +ENQN N
Sbjct: 96 -------SSNNKDTYYYNKNAYESTKQQNLGEAIFTEKG--WSTKENQNNNY-------- 138
Query: 577 AANENYNFNNNNAANNNEKGAEVQGMSDTRFMEGGKYFYNVNSENEKYNPTGYGGESSTR 398
+N NN NN EK QGMSDTR++E GKY+Y+V SEN Y P + ++R
Sbjct: 139 -------YNGNNGYNNGEK----QGMSDTRYLENGKYYYDVKSEN-NYYPNRF---DNSR 183
Query: 397 GVNSENWYNNKGYFGNNNNENKNSMEGY-KNQEEFEDDPDVDFEP 266
GV S N +N Y N+M Y +NQEEFE+ + +FEP
Sbjct: 184 GVASRNEFNENRY---------NNMGRYHQNQEEFEESEE-EFEP 218
>pir||S65063 fiber protein E6 (clones SIE6-2A and SIE6-3B) - sea-island cotton
gi|1000088|gb|AAB03081.1| E6 gi|1000090|gb|AAB03085.1| E6
Length = 246
Score = 136 bits (342), Expect = 7e-31
Identities = 111/300 (37%), Positives = 147/300 (49%), Gaps = 6/300 (2%)
Frame = -1
Query: 1147 LKILFFLLLNNPVDCPMQINARDSQFFSKVTHVNNNNNNVKETEFPNNNEVP--VNKPEQ 974
+ ILF L + MQI+AR+ +FSK VN N E + VP KPE+
Sbjct: 10 MSILFLFALFS-----MQIHARE--YFSKFPRVNINEKETTTREQKHETFVPQTTQKPEE 62
Query: 973 Q-PVFTPETENSYGLYGHESGLHPPTTTTTYLP-YKTTPSKEDHTTTKNFNNNYNNYQKD 800
Q P F PET+N YGLYGHESG + + + P + T + E + T F+ D
Sbjct: 63 QEPRFIPETQNGYGLYGHESGSGSGSGSGSSRPSFTTKETYEPYVTPVRFH-------PD 115
Query: 799 DFFNTNQNDQLSDTRFIGNSHSSSNNNNYYYNNKNAYEGN-QNELSNTKFTEGGNKYNPR 623
+ +N+ SSNN + YY NKNAYE Q L FTE G ++ +
Sbjct: 116 EPYNSIPE--------------SSNNKDTYYYNKNAYESTKQQNLGEAIFTEKG--WSTK 159
Query: 622 ENQNYNQKFFYNNNAAANENYNFNNNNAANNNEKGAEVQGMSDTRFMEGGKYFYNVNSEN 443
ENQN N +N NN NN EK QGMSDTR++E GKY+Y+V SEN
Sbjct: 160 ENQNNNY---------------YNGNNGYNNGEK----QGMSDTRYLENGKYYYDVKSEN 200
Query: 442 EKYNPTGYGGESSTRGVNSENWYNNKGYFGNNNNENKNSMEGY-KNQEEFEDDPDVDFEP 266
Y P + ++RGV S N +N Y N+M Y +NQEEFE+ + +FEP
Sbjct: 201 -NYYPNRF---DNSRGVASRNEFNENRY---------NNMGRYHQNQEEFEESEE-EFEP 246
>pir||S65062 fiber protein E6 (clone CKE6-4A) - upland cotton
gi|1000086|gb|AAB03080.1| E6
gi|9651644|gb|AAF91226.1|AF218378_1 protein kinase
[Gossypium hirsutum]
Length = 241
Score = 126 bits (317), Expect = 6e-28
Identities = 109/307 (35%), Positives = 140/307 (45%), Gaps = 13/307 (4%)
Frame = -1
Query: 1147 LKILFFLLLNNPVDCPMQINARDSQFFSKVTHVNNNNNNVKETEFPNNNEVP--VNKPEQ 974
+ ILF L + MQI+AR+ +FSK VN N E + VP KPE+
Sbjct: 10 MSILFLFALFS-----MQIHARE--YFSKFPRVNTNEKETTTREQEHETFVPQTTQKPEE 62
Query: 973 Q-PVFTPETENSYGLYGHESGLHPP--------TTTTTYLPYKTTPSKEDHTTTKNFNNN 821
Q P F PET+N YGLYGHESG TT TY PY TP +
Sbjct: 63 QEPRFIPETQNGYGLYGHESGSGSGSGSSRPSFTTKETYEPY-VTPVR------------ 109
Query: 820 YNNYQKDDFFNTNQNDQLSDTRFIGNSHSSSNNNNYYYNNKNAYEGN-QNELSNTKFTEG 644
+ D+ +N+ SSNN + YY NKNAY+ Q L FTE
Sbjct: 110 ---FHPDEPYNSIPE--------------SSNNKDTYYYNKNAYKSTKQQNLGEAIFTEK 152
Query: 643 GNKYNPRENQNYNQKFFYNNNAAANENYNFNNNNAANNNEKGAEVQGMSDTRFMEGGKYF 464
G ++ +ENQN N +YN N N EK QGMSDTR++E GKY+
Sbjct: 153 G--WSTKENQNNN---YYNGNI---------------NGEK----QGMSDTRYLENGKYY 188
Query: 463 YNVNSENEKY-NPTGYGGESSTRGVNSENWYNNKGYFGNNNNENKNSMEGYKNQEEFEDD 287
Y+V SEN Y N ++R EN YNN G + ++NQEEFE+
Sbjct: 189 YDVKSENSYYPNQLDNSRGVASRNEFDENRYNNMGRY-------------HQNQEEFEES 235
Query: 286 PDVDFEP 266
+ +FEP
Sbjct: 236 EE-EFEP 241
>pir||T10265 arabinogalactan-protein AGP2 - Persian tobacco
gi|1087017|gb|AAB35284.1| arabinogalactan-protein; AGP
[Nicotiana alata]
Length = 461
Score = 114 bits (285), Expect = 3e-24
Identities = 85/273 (31%), Positives = 126/273 (46%), Gaps = 17/273 (6%)
Frame = -1
Query: 1048 NNNNNNVKETEFPNNNEVPVNKPEQQPVFTPETENSYGLYGHESGLHPPTTTTTYLPYKT 869
NNNNN+ +E NNN N + E N+ G+ + Y
Sbjct: 196 NNNNNDDGFSENYNNNGYSENANNKNNNGYSENYNNNNNNGYAKNYNNG--------YSQ 247
Query: 868 TPSKEDHTTTKNFNNNYNNYQKDDFFNTNQNDQLSDTRFIGNSHSSSNNNNYY---YNNK 698
+ + ++ ++N+NNN NN + N+N N G S + NNNN + YNN
Sbjct: 248 SYNNNNNFYSENYNNNNNNVFSE---NSNNNGYSKKINNNGYSQNYMNNNNGFSESYNNN 304
Query: 697 NAYEGNQNELS-NTKFTEGGNKYNPRENQNYNQKFFYNNNAAANENYNFNNNNAA--NNN 527
N N N S N N ++ N N N FY N N Y+ N N A+ NNN
Sbjct: 305 NNNNNNNNVFSENYNNNNNNNVFSENYNNNNNNNAFYENYNNNNNGYSENYNQASSYNNN 364
Query: 526 EKGAEVQGMSDTRFMEGGKYFYNVNSEN-------EKYN-PTGYGGESS---TRGVNSEN 380
+ E QG+SDTRF+E GKY+Y++ +EN E YN + Y ++ +G++
Sbjct: 365 DNTVERQGLSDTRFLENGKYYYDIKNENTNNNGYSENYNHVSSYNNNNNMVERQGLSDTR 424
Query: 379 WYNNKGYFGNNNNENKNSMEGYKNQEEFEDDPD 281
+ +N YF +NN E K S+E + Q+E+ D D
Sbjct: 425 FLDNGNYFYSNNGE-KMSVEESERQQEYPDTED 456
Score = 82.8 bits (203), Expect = 9e-15
Identities = 70/253 (27%), Positives = 95/253 (36%), Gaps = 8/253 (3%)
Frame = -1
Query: 982 PEQQPVFTPE---TENSYGLYGHESGLHPPTTTTTYLPYKTTPSKEDHTTTKNFNNNYNN 812
PE+ + P T+ YGLYG S T T TP+KE N + +YNN
Sbjct: 118 PEEGGIEAPAPLLTDTPYGLYGPHSQEISSTVTNLDEVETQTPAKEFQGARFNTDESYNN 177
Query: 811 YQKDDFFNTNQNDQLSDTRFIGNSHSSSNNNNYYYNNKNAYEGNQNELSNTKFTEG---- 644
D N N N+ D+ N++ + NY N N Y N N +N ++E
Sbjct: 178 NGYDS--NNNDNNNGYDSNNNNNNNDDGFSENY---NNNGYSENANNKNNNGYSENYNNN 232
Query: 643 -GNKYNPRENQNYNQKFFYNNNAAANENYNFNNNNAANNNEKGAEVQGMSDTRFMEGGKY 467
N Y N Y+Q + NNN +ENYN NNNN + N
Sbjct: 233 NNNGYAKNYNNGYSQSY-NNNNNFYSENYNNNNNNVFSENS------------------- 272
Query: 466 FYNVNSENEKYNPTGYGGESSTRGVNSENWYNNKGYFGNNNNENKNSMEGYKNQEEFEDD 287
N N ++K N GY YNN NNNN + N F ++
Sbjct: 273 --NNNGYSKKINNNGYSQNYMNNNNGFSESYNNNNNNNNNNNVFSENYNNNNNNNVFSEN 330
Query: 286 PDVDFEP*AFNHN 248
+ + AF N
Sbjct: 331 YNNNNNNNAFYEN 343
Score = 60.8 bits (146), Expect = 4e-08
Identities = 62/226 (27%), Positives = 82/226 (35%), Gaps = 8/226 (3%)
Frame = -1
Query: 1099 MQINARDSQFFSKVTHVNNNNNNVKETEFPNNNEVPVNKPEQQPVFTPETENSYGLYGHE 920
M N S+ ++ + NNNNN E NNN VF+ N
Sbjct: 291 MNNNNGFSESYNNNNNNNNNNNVFSENYNNNNN---------NNVFSENYNN-------- 333
Query: 919 SGLHPPTTTTTYLPYKTTPSKEDHTTTKNFNNNYN----NYQKDDFFNTNQN----DQLS 764
+ ++ +N+NNN N NY + +N N N LS
Sbjct: 334 -------------------NNNNNAFYENYNNNNNGYSENYNQASSYNNNDNTVERQGLS 374
Query: 763 DTRFIGNSHSSSNNNNYYYNNKNAYEGNQNELSNTKFTEGGNKYNPRENQNYNQKFFYNN 584
DTRF+ N YYY+ KN N N ++NYN YN
Sbjct: 375 DTRFL-------ENGKYYYDIKNE-----------------NTNNNGYSENYNHVSSYN- 409
Query: 583 NAAANENYNFNNNNAANNNEKGAEVQGMSDTRFMEGGKYFYNVNSE 446
NNNN E QG+SDTRF++ G YFY+ N E
Sbjct: 410 ----------NNNNM-------VERQGLSDTRFLDNGNYFYSNNGE 438
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,017,442,010
Number of Sequences: 1393205
Number of extensions: 26770213
Number of successful extensions: 626969
Number of sequences better than 10.0: 6097
Number of HSP's better than 10.0 without gapping: 105808
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 239531
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 71654580342
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)