Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
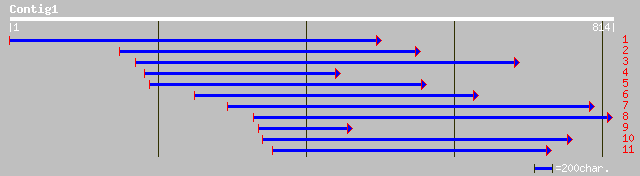
Query= KMC002921A_C01 KMC002921A_c01
(808 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_182067.1| putative AT-hook DNA-binding protein; protein i... 132 5e-39
ref|NP_191646.1| putative protein; protein id: At3g60870.1 [Arab... 127 3e-34
ref|NP_192942.1| putative DNA-binding protein; protein id: At4g1... 142 6e-33
ref|NP_194012.1| putative DNA binding protein; protein id: At4g2... 135 9e-31
ref|NP_173514.1| putative DNA-binding protein; protein id: At1g2... 120 2e-26
>ref|NP_182067.1| putative AT-hook DNA-binding protein; protein id: At2g45430.1
[Arabidopsis thaliana] gi|25348246|pir||D84890 probable
AT-hook DNA-binding protein [imported] - Arabidopsis
thaliana gi|2583112|gb|AAB82621.1| putative AT-hook
DNA-binding protein [Arabidopsis thaliana]
Length = 317
Score = 132 bits (332), Expect(2) = 5e-39
Identities = 65/77 (84%), Positives = 70/77 (90%)
Frame = -2
Query: 807 FEILSLSGSFLPPPAPPAASGLAIYLAGGQGQVVGGSVVGPLLASGPVVIMAASFGNAAY 628
FEILSLSGSFLPPPAPPAASGL IYLAGGQGQVVGGSVVGPL+ASGPVVIMAASFGNAAY
Sbjct: 178 FEILSLSGSFLPPPAPPAASGLTIYLAGGQGQVVGGSVVGPLMASGPVVIMAASFGNAAY 237
Query: 627 ERLPLEDEETPATVTGS 577
ERLPLE+++ G+
Sbjct: 238 ERLPLEEDDQEEQTAGA 254
Score = 51.6 bits (122), Expect(2) = 5e-39
Identities = 27/48 (56%), Positives = 31/48 (64%)
Frame = -3
Query: 536 QQPQSQPQQQQLMPDPNASLFHGVPQNLLNSCQLPSEGYWNGTCSPTF 393
Q Q QQQQLM DP S G+P NL+NS QLP+E YW GT P+F
Sbjct: 272 QTQTQQQQQQQLMQDPT-SFIQGLPPNLMNSVQLPAEAYW-GTPRPSF 317
>ref|NP_191646.1| putative protein; protein id: At3g60870.1 [Arabidopsis thaliana]
gi|11281680|pir||T47898 hypothetical protein T4C21.280 -
Arabidopsis thaliana gi|7329697|emb|CAB82691.1| putative
protein [Arabidopsis thaliana]
Length = 265
Score = 127 bits (319), Expect(2) = 3e-34
Identities = 61/69 (88%), Positives = 66/69 (95%)
Frame = -2
Query: 807 FEILSLSGSFLPPPAPPAASGLAIYLAGGQGQVVGGSVVGPLLASGPVVIMAASFGNAAY 628
FEILSLSGSFLPPPAPPAASGL +YLAGGQGQV+GGSVVGPL AS PVV+MAASFGNA+Y
Sbjct: 142 FEILSLSGSFLPPPAPPAASGLKVYLAGGQGQVIGGSVVGPLTASSPVVVMAASFGNASY 201
Query: 627 ERLPLEDEE 601
ERLPLE+EE
Sbjct: 202 ERLPLEEEE 210
Score = 40.4 bits (93), Expect(2) = 3e-34
Identities = 22/40 (55%), Positives = 26/40 (65%)
Frame = -3
Query: 512 QQQLMPDPNASLFHGVPQNLLNSCQLPSEGYWNGTCSPTF 393
Q+QLM D A+ F G P NL+NS LP E YW GT P+F
Sbjct: 229 QKQLMQD--ATSFIGSPSNLINSVSLPGEAYW-GTQRPSF 265
>ref|NP_192942.1| putative DNA-binding protein; protein id: At4g12050.1 [Arabidopsis
thaliana] gi|7485568|pir||T06612 hypothetical protein
F16J13.120 - Arabidopsis thaliana
gi|4586110|emb|CAB40946.1| putative DNA-binding protein
[Arabidopsis thaliana] gi|7267906|emb|CAB78248.1|
putative DNA-binding protein [Arabidopsis thaliana]
Length = 339
Score = 142 bits (358), Expect = 6e-33
Identities = 73/103 (70%), Positives = 82/103 (78%), Gaps = 7/103 (6%)
Frame = -2
Query: 807 FEILSLSGSFLPPPAPPAASGLAIYLAGGQGQVVGGSVVGPLLASGPVVIMAASFGNAAY 628
FEILSLSGSFLPPPAPPAA+GL++YLAGGQGQVVGGSVVGPLL SGPVV+MAASF NAAY
Sbjct: 204 FEILSLSGSFLPPPAPPAATGLSVYLAGGQGQVVGGSVVGPLLCSGPVVVMAASFSNAAY 263
Query: 627 ERLPLEDEETPATVT-------GSGGLGSPGIMGTAAATTAIA 520
ERLPLE++E V G GG+GSP +MG A A+A
Sbjct: 264 ERLPLEEDEMQTPVQGGGGGGGGGGGMGSPPMMGQQQAMAAMA 306
>ref|NP_194012.1| putative DNA binding protein; protein id: At4g22810.1 [Arabidopsis
thaliana] gi|7486887|pir||T04576 hypothetical protein
T12H17.200 - Arabidopsis thaliana
gi|2827558|emb|CAA16566.1| putative DNA binding protein
[Arabidopsis thaliana] gi|7269128|emb|CAB79236.1|
putative DNA binding protein [Arabidopsis thaliana]
Length = 324
Score = 135 bits (339), Expect = 9e-31
Identities = 68/90 (75%), Positives = 76/90 (83%), Gaps = 3/90 (3%)
Frame = -2
Query: 807 FEILSLSGSFLPPPAPPAASGLAIYLAGGQGQVVGGSVVGPLLASGPVVIMAASFGNAAY 628
FEILSLSGSFLPPPAPP A+GL++YLAGGQGQVVGGSVVGPLL +GPVV+MAASF NAAY
Sbjct: 193 FEILSLSGSFLPPPAPPTATGLSVYLAGGQGQVVGGSVVGPLLCAGPVVVMAASFSNAAY 252
Query: 627 ERLPLEDEETPATVTGSGG---LGSPGIMG 547
ERLPLE++E V G GG L SP +MG
Sbjct: 253 ERLPLEEDEMQTPVHGGGGGGSLESPPMMG 282
>ref|NP_173514.1| putative DNA-binding protein; protein id: At1g20900.1, supported by
cDNA: gi_6319179 [Arabidopsis thaliana]
gi|20532086|sp|Q9S7C9|ESCA_ARATH Putative DNA-binding
protein ESCAROLA gi|25348247|pir||F86341 hypothetical
protein F9H16.12 - Arabidopsis thaliana
gi|4836899|gb|AAD30602.1|AC007369_12 Unknown protein
[Arabidopsis thaliana]
gi|6319180|gb|AAF07197.1|AF194974_1 ESCAROLA
[Arabidopsis thaliana]
Length = 311
Score = 120 bits (302), Expect = 2e-26
Identities = 61/105 (58%), Positives = 74/105 (70%)
Frame = -2
Query: 807 FEILSLSGSFLPPPAPPAASGLAIYLAGGQGQVVGGSVVGPLLASGPVVIMAASFGNAAY 628
FEILSL+G+ LPPPAPP A GL+I+LAGGQGQVVGGSVV PL+AS PV++MAASF NA +
Sbjct: 180 FEILSLTGTVLPPPAPPGAGGLSIFLAGGQGQVVGGSVVAPLIASAPVILMAASFSNAVF 239
Query: 627 ERLPLEDEETPATVTGSGGLGSPGIMGTAAATTAIATTTATTHAG 493
ERLP+E+EE G GG G P M A + + + T G
Sbjct: 240 ERLPIEEEEEEGGGGGGGGGGGPPQMQQAPSASPPSGVTGQGQLG 284
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 764,769,139
Number of Sequences: 1393205
Number of extensions: 19242923
Number of successful extensions: 198490
Number of sequences better than 10.0: 262
Number of HSP's better than 10.0 without gapping: 88341
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 164924
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 41176381974
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)