Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002915A_C01 KMC002915A_c01
(683 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
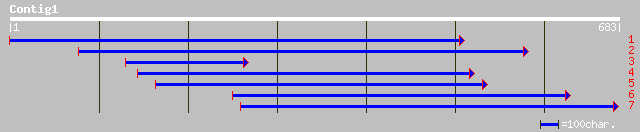
ref|NP_177591.1| hypothetical protein; protein id: At1g74510.1 [... 83 4e-15
ref|NP_178390.1| predicted by genefinder and genscan; protein id... 70 2e-11
gb|AAM98120.1| predicted protein [Arabidopsis thaliana] 70 2e-11
ref|NP_174015.1| unknown protein; protein id: At1g26930.1 [Arabi... 70 2e-11
ref|NP_172885.1| hypothetical protein; protein id: At1g14330.1, ... 68 1e-10
>ref|NP_177591.1| hypothetical protein; protein id: At1g74510.1 [Arabidopsis
thaliana] gi|25406351|pir||B96774 hypothetical protein
F1M20.19 [imported] - Arabidopsis thaliana
gi|12324791|gb|AAG52353.1|AC011765_5 hypothetical
protein; 62385-63740 [Arabidopsis thaliana]
gi|28973619|gb|AAO64134.1| unknown protein [Arabidopsis
thaliana]
Length = 451
Score = 82.8 bits (203), Expect = 4e-15
Identities = 36/55 (65%), Positives = 46/55 (83%), Gaps = 1/55 (1%)
Frame = -1
Query: 683 ACGNRLIVIGGPRALDGRVIEVNACVPGEG-EPEWNLLASRQSGSFVYNCAVMGC 522
ACG++L+V+GGPRA+ G IE+NACVP EG + W +LAS+ SG+FVYNCAVMGC
Sbjct: 397 ACGDQLVVVGGPRAIGGGFIEINACVPSEGTQLHWRVLASKPSGNFVYNCAVMGC 451
>ref|NP_178390.1| predicted by genefinder and genscan; protein id: At2g02870.1,
supported by cDNA: gi_16974559 [Arabidopsis thaliana]
gi|25410988|pir||H84441 hypothetical protein At2g02870
[imported] - Arabidopsis thaliana
gi|3461814|gb|AAC32908.1| predicted by genefinder and
genscan [Arabidopsis thaliana]
gi|16974560|gb|AAL31196.1| At2g02870/T17M13.4
[Arabidopsis thaliana] gi|25090100|gb|AAN72228.1|
At2g02870/T17M13.4 [Arabidopsis thaliana]
Length = 467
Score = 70.5 bits (171), Expect = 2e-11
Identities = 32/55 (58%), Positives = 40/55 (72%), Gaps = 1/55 (1%)
Frame = -1
Query: 683 ACGNRLIVIGGPRALDGRVIEVNACVPGE-GEPEWNLLASRQSGSFVYNCAVMGC 522
ACG RLIVIGGP+ G IE+N+ +P + G P+W LL + S +FVYNCAVMGC
Sbjct: 413 ACGERLIVIGGPKCSGGGFIELNSWIPSDGGPPQWTLLDRKHSPTFVYNCAVMGC 467
>gb|AAM98120.1| predicted protein [Arabidopsis thaliana]
Length = 467
Score = 70.5 bits (171), Expect = 2e-11
Identities = 32/55 (58%), Positives = 40/55 (72%), Gaps = 1/55 (1%)
Frame = -1
Query: 683 ACGNRLIVIGGPRALDGRVIEVNACVPGE-GEPEWNLLASRQSGSFVYNCAVMGC 522
ACG RLIVIGGP+ G IE+N+ +P + G P+W LL + S +FVYNCAVMGC
Sbjct: 413 ACGERLIVIGGPKCSGGGFIELNSWIPSDGGPPQWTLLDRKHSPTFVYNCAVMGC 467
>ref|NP_174015.1| unknown protein; protein id: At1g26930.1 [Arabidopsis thaliana]
gi|25518438|pir||C86396 hypothetical protein T2P11.12
[imported] - Arabidopsis thaliana
gi|4262182|gb|AAD14499.1| 44123
Length = 404
Score = 70.5 bits (171), Expect = 2e-11
Identities = 32/54 (59%), Positives = 40/54 (73%)
Frame = -1
Query: 683 ACGNRLIVIGGPRALDGRVIEVNACVPGEGEPEWNLLASRQSGSFVYNCAVMGC 522
ACG+R+IVIGGP+A IE+N+ VP PEW+LL +QS +FVYNCAVM C
Sbjct: 351 ACGDRIIVIGGPKAPGEGFIELNSWVPSVTTPEWHLLGKKQSVNFVYNCAVMSC 404
>ref|NP_172885.1| hypothetical protein; protein id: At1g14330.1, supported by cDNA:
gi_20453204 [Arabidopsis thaliana]
gi|25513629|pir||F86277 F14L17.10 protein - Arabidopsis
thaliana gi|7262675|gb|AAF43933.1|AC012188_10 Contains
strong similarity to a hypothetical protein from
Arabidopsis thaliana gb|AC004138.2 and contains three
Kelch PF|01344 domains. EST gb|Z26791 comes from this
gene gi|20453205|gb|AAM19842.1| At1g14330/F14L17_7
[Arabidopsis thaliana] gi|23308401|gb|AAN18170.1|
At1g14330/F14L17_7 [Arabidopsis thaliana]
Length = 441
Score = 68.2 bits (165), Expect = 1e-10
Identities = 32/56 (57%), Positives = 38/56 (67%), Gaps = 2/56 (3%)
Frame = -1
Query: 683 ACGNRLIVIGGPRALDGRVIEVNACVPG--EGEPEWNLLASRQSGSFVYNCAVMGC 522
ACG RLIVIGGPR+ G IE+N+ +P P W LL + S +FVYNCAVMGC
Sbjct: 386 ACGERLIVIGGPRSSGGGYIELNSWIPSSDRSPPLWTLLGRKHSSNFVYNCAVMGC 441
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 565,502,760
Number of Sequences: 1393205
Number of extensions: 11888273
Number of successful extensions: 26179
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 25342
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26165
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30552968016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)