Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002909A_C01 KMC002909A_c01
(958 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
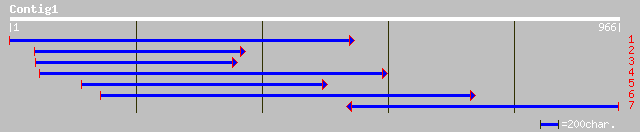
pir||F84631 hypothetical protein At2g24020 [imported] - Arabidop... 212 6e-54
ref|NP_565561.1| expressed protein; protein id: At2g24020.1, sup... 212 6e-54
gb|AAM62959.1| unknown [Arabidopsis thaliana] 211 1e-53
ref|NP_194791.1| putative protein; protein id: At4g30620.1, supp... 211 1e-53
gb|AAO32624.1| CR084 protein [Chlamydomonas reinhardtii] 118 1e-25
>pir||F84631 hypothetical protein At2g24020 [imported] - Arabidopsis thaliana
Length = 179
Score = 212 bits (540), Expect = 6e-54
Identities = 116/162 (71%), Positives = 128/162 (78%), Gaps = 3/162 (1%)
Frame = -3
Query: 821 LNSSANVVDMRPLSQRGHKKVGRDRCFRVYALFGG-KKENN--DGDSKTGMLGNMQNLYE 651
++ +A++ R + +K R RV LFGG K+NN DG SK G+ GNMQN+YE
Sbjct: 18 VSGNASLNSQRRTWPKQYKSKNGYRSLRVNGLFGGGNKDNNSEDGQSKAGIFGNMQNMYE 77
Query: 650 TVKKAQMVVQVEAVRVQKELAAAEFDGYCEDELIKVTLSGNQQPIRTIITEAAMELGPEK 471
TVKKAQMVVQVEAVRVQKELAAAEFDGYC EL+KVTLSGNQQPIRT ITEAAMELG EK
Sbjct: 78 TVKKAQMVVQVEAVRVQKELAAAEFDGYCAGELVKVTLSGNQQPIRTDITEAAMELGSEK 137
Query: 470 LSLLVTEAYKDAHQKSVQAMKERMSGLAQSLGMPPGLGEGLK 345
LS LVTEAYKDAH KSV AMKERMS LAQSLGMPPGL EG+K
Sbjct: 138 LSQLVTEAYKDAHAKSVVAMKERMSDLAQSLGMPPGLSEGMK 179
>ref|NP_565561.1| expressed protein; protein id: At2g24020.1, supported by cDNA:
gi_15081683, supported by cDNA: gi_20147168 [Arabidopsis
thaliana] gi|15081684|gb|AAK82497.1| At2g24020/T29E15.22
[Arabidopsis thaliana] gi|20147169|gb|AAM10301.1|
At2g24020/T29E15.22 [Arabidopsis thaliana]
gi|20197299|gb|AAC63670.2| expressed protein
[Arabidopsis thaliana]
Length = 182
Score = 212 bits (540), Expect = 6e-54
Identities = 116/162 (71%), Positives = 128/162 (78%), Gaps = 3/162 (1%)
Frame = -3
Query: 821 LNSSANVVDMRPLSQRGHKKVGRDRCFRVYALFGG-KKENN--DGDSKTGMLGNMQNLYE 651
++ +A++ R + +K R RV LFGG K+NN DG SK G+ GNMQN+YE
Sbjct: 21 VSGNASLNSQRRTWPKQYKSKNGYRSLRVNGLFGGGNKDNNSEDGQSKAGIFGNMQNMYE 80
Query: 650 TVKKAQMVVQVEAVRVQKELAAAEFDGYCEDELIKVTLSGNQQPIRTIITEAAMELGPEK 471
TVKKAQMVVQVEAVRVQKELAAAEFDGYC EL+KVTLSGNQQPIRT ITEAAMELG EK
Sbjct: 81 TVKKAQMVVQVEAVRVQKELAAAEFDGYCAGELVKVTLSGNQQPIRTDITEAAMELGSEK 140
Query: 470 LSLLVTEAYKDAHQKSVQAMKERMSGLAQSLGMPPGLGEGLK 345
LS LVTEAYKDAH KSV AMKERMS LAQSLGMPPGL EG+K
Sbjct: 141 LSQLVTEAYKDAHAKSVVAMKERMSDLAQSLGMPPGLSEGMK 182
>gb|AAM62959.1| unknown [Arabidopsis thaliana]
Length = 180
Score = 211 bits (537), Expect = 1e-53
Identities = 113/138 (81%), Positives = 121/138 (86%), Gaps = 2/138 (1%)
Frame = -3
Query: 752 DRCFRVYALFGGKKENN--DGDSKTGMLGNMQNLYETVKKAQMVVQVEAVRVQKELAAAE 579
+R RV LFGG K++N DG SK G+LGNMQNLYETVKKAQMVVQVEAVRVQKELA AE
Sbjct: 44 NRSLRVNVLFGGGKKDNKEDGQSKAGILGNMQNLYETVKKAQMVVQVEAVRVQKELAVAE 103
Query: 578 FDGYCEDELIKVTLSGNQQPIRTIITEAAMELGPEKLSLLVTEAYKDAHQKSVQAMKERM 399
FDGYC+ EL+KVTLSGNQQPIRT IT+AAMELG EKLSLLVTEAYKDAH KSV AMKERM
Sbjct: 104 FDGYCQGELVKVTLSGNQQPIRTDITDAAMELGSEKLSLLVTEAYKDAHSKSVLAMKERM 163
Query: 398 SGLAQSLGMPPGLGEGLK 345
S LAQSLGMPPGL +GLK
Sbjct: 164 SDLAQSLGMPPGL-DGLK 180
>ref|NP_194791.1| putative protein; protein id: At4g30620.1, supported by cDNA:
17578., supported by cDNA: gi_18650640 [Arabidopsis
thaliana] gi|25407667|pir||C85358 hypothetical protein
AT4g30620 [imported] - Arabidopsis thaliana
gi|7269963|emb|CAB79780.1| putative protein [Arabidopsis
thaliana] gi|18650641|gb|AAL75890.1| AT4g30620/F17I23_40
[Arabidopsis thaliana]
Length = 180
Score = 211 bits (537), Expect = 1e-53
Identities = 113/138 (81%), Positives = 121/138 (86%), Gaps = 2/138 (1%)
Frame = -3
Query: 752 DRCFRVYALFGGKKENN--DGDSKTGMLGNMQNLYETVKKAQMVVQVEAVRVQKELAAAE 579
+R RV LFGG K++N DG SK G+LGNMQNLYETVKKAQMVVQVEAVRVQKELA AE
Sbjct: 44 NRSLRVNGLFGGGKKDNKEDGQSKAGILGNMQNLYETVKKAQMVVQVEAVRVQKELAVAE 103
Query: 578 FDGYCEDELIKVTLSGNQQPIRTIITEAAMELGPEKLSLLVTEAYKDAHQKSVQAMKERM 399
FDGYC+ EL+KVTLSGNQQPIRT IT+AAMELG EKLSLLVTEAYKDAH KSV AMKERM
Sbjct: 104 FDGYCQGELVKVTLSGNQQPIRTDITDAAMELGSEKLSLLVTEAYKDAHSKSVLAMKERM 163
Query: 398 SGLAQSLGMPPGLGEGLK 345
S LAQSLGMPPGL +GLK
Sbjct: 164 SDLAQSLGMPPGL-DGLK 180
>gb|AAO32624.1| CR084 protein [Chlamydomonas reinhardtii]
Length = 157
Score = 118 bits (296), Expect = 1e-25
Identities = 66/127 (51%), Positives = 88/127 (68%), Gaps = 1/127 (0%)
Frame = -3
Query: 740 RVYALFGGKKENNDGDSKTGMLGNMQNLYETVKKAQMVVQVEAVRVQKELAAAEFDGYCE 561
+V ALFGG G G +M+NL E+VKKAQ +VQ E RVQ ELAA EF+GY E
Sbjct: 33 QVKALFGG------GQGGGGNPFDMKNLMESVKKAQQLVQTETARVQAELAATEFEGYDE 86
Query: 560 DELIKVTLSGNQQPIRTIITEAAMELGPEKLSLLVTEAYKDAHQKSVQAMKERMSGLAQS 381
+E ++V +SGNQ+P IT+AA++LG E+ S T+A +DAH+KSV MKE+M LA++
Sbjct: 87 EETVRVIMSGNQEPKGVEITQAALDLGAEECSKRTTDAMRDAHKKSVTGMKEKMRELAKN 146
Query: 380 LGMP-PG 363
LG+P PG
Sbjct: 147 LGIPNPG 153
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 805,334,485
Number of Sequences: 1393205
Number of extensions: 16595291
Number of successful extensions: 41996
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 40339
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 41867
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 54078381240
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)