Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002901A_C01 KMC002901A_c01
(796 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
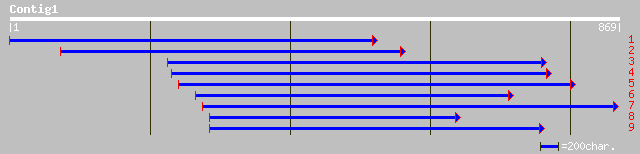
gb|AAM61252.1| unknown [Arabidopsis thaliana] 69 8e-11
ref|NP_567964.1| expressed protein; protein id: At4g34340.1, sup... 67 4e-10
dbj|BAA92559.1| KIAA1321 protein [Homo sapiens] 38 0.19
gb|EAA12317.1| agCP11454 [Anopheles gambiae str. PEST] 37 0.33
ref|NP_648004.1| CG13290-PA [Drosophila melanogaster] gi|7295407... 37 0.33
>gb|AAM61252.1| unknown [Arabidopsis thaliana]
Length = 353
Score = 68.9 bits (167), Expect = 8e-11
Identities = 43/100 (43%), Positives = 57/100 (57%)
Frame = -1
Query: 793 VSVLEAFAPAIEVLGSSSLCDDDGVEGRMVVPAARPTVHFKFRTGKKLIGESLDARLQKK 614
+SV+EAFAPA+E D E +P K RT KK +G+ LD LQKK
Sbjct: 261 LSVIEAFAPAMEAAR-----DGFSSEAHTEWKKNKPVALSKLRTEKKFLGQPLDLSLQKK 315
Query: 613 DAASRAASLGGREDERDDKKRRAEYILRQSIENPQELTLL 494
+ + RE++RDDK+RRAE+ILRQ +ENP +L L
Sbjct: 316 GEDRPISFV--REEDRDDKRRRAEFILRQCMENPVDLNQL 353
>ref|NP_567964.1| expressed protein; protein id: At4g34340.1, supported by cDNA:
115167., supported by cDNA: gi_13430531 [Arabidopsis
thaliana] gi|7485312|pir||T04780 hypothetical protein
F10M10.110 - Arabidopsis thaliana
gi|4455179|emb|CAB36711.1| hypothetical protein
[Arabidopsis thaliana] gi|7270384|emb|CAB80151.1|
hypothetical protein [Arabidopsis thaliana]
gi|13430532|gb|AAK25888.1|AF360178_1 unknown protein
[Arabidopsis thaliana] gi|14532740|gb|AAK64071.1|
unknown protein [Arabidopsis thaliana]
Length = 353
Score = 66.6 bits (161), Expect = 4e-10
Identities = 42/100 (42%), Positives = 56/100 (56%)
Frame = -1
Query: 793 VSVLEAFAPAIEVLGSSSLCDDDGVEGRMVVPAARPTVHFKFRTGKKLIGESLDARLQKK 614
+SV+EAFAPA+E D E +P K RT KK +G+ LD LQ K
Sbjct: 261 LSVIEAFAPAMEAAR-----DGFSSEAHTEWKKNKPVALSKLRTEKKFLGQPLDLSLQMK 315
Query: 613 DAASRAASLGGREDERDDKKRRAEYILRQSIENPQELTLL 494
+ + RE++RDDK+RRAE+ILRQ +ENP +L L
Sbjct: 316 GEDRPISFV--REEDRDDKRRRAEFILRQCMENPVDLNQL 353
>dbj|BAA92559.1| KIAA1321 protein [Homo sapiens]
Length = 714
Score = 37.7 bits (86), Expect = 0.19
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 1/40 (2%)
Frame = +1
Query: 679 AQSASQQEPPFSPQPHHHHTNYY-SLEPQSQEQKPPTQKH 795
+Q S +E P PQP HHH++++ PQ Q+Q+P H
Sbjct: 15 SQPLSMEEKPGQPQPQHHHSHHHPHHHPQQQQQQPHHHHH 54
>gb|EAA12317.1| agCP11454 [Anopheles gambiae str. PEST]
Length = 268
Score = 37.0 bits (84), Expect = 0.33
Identities = 20/55 (36%), Positives = 30/55 (54%), Gaps = 4/55 (7%)
Frame = +1
Query: 637 KIPQ*ASSQF*T*NAQSAS----QQEPPFSPQPHHHHTNYYSLEPQSQEQKPPTQ 789
++P +SS T A +AS QQ+ PQPHHHH ++ Q Q+Q+ +Q
Sbjct: 19 RLPHQSSSSSSTTAAVNASSSPHQQQQQQQPQPHHHHAQQQHMQQQQQQQQQHSQ 73
Score = 35.8 bits (81), Expect = 0.74
Identities = 12/26 (46%), Positives = 18/26 (69%)
Frame = +1
Query: 682 QSASQQEPPFSPQPHHHHTNYYSLEP 759
Q QQ+PP +P PHHHH ++++ P
Sbjct: 78 QHQQQQQPPSAPPPHHHHHHHHAHHP 103
>ref|NP_648004.1| CG13290-PA [Drosophila melanogaster] gi|7295407|gb|AAF50724.1|
CG13290-PA [Drosophila melanogaster]
Length = 169
Score = 37.0 bits (84), Expect = 0.33
Identities = 27/90 (30%), Positives = 38/90 (42%), Gaps = 7/90 (7%)
Frame = +1
Query: 541 TQPSSSYHPSHLLCHPTTPLAKQHPSSEV---*HPKIPQ*ASSQF*T*NA----QSASQQ 699
T P +H H HP P + HP S HP++ + Q ++ Q S Q
Sbjct: 57 TSPQHFFHHHHPPAHPHPPRQQPHPHSHSHPHPHPQLQRRPVEQLHLLHSHHDVQELSGQ 116
Query: 700 EPPFSPQPHHHHTNYYSLEPQSQEQKPPTQ 789
E P PQP H ++ P S+E PT+
Sbjct: 117 EHP-HPQPGSHPHPHHLRSPSSEEDNSPTE 145
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 640,249,926
Number of Sequences: 1393205
Number of extensions: 13275355
Number of successful extensions: 45625
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 40511
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 44702
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40055936206
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)