Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002892A_C01 KMC002892A_c01
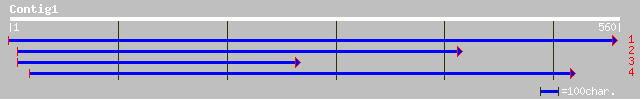
(555 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91628.1| unknown protein [Arabidopsis thaliana] 87 2e-16
ref|NP_193198.1| hypothetical protein; protein id: At4g14620.1 [... 87 2e-16
pir||G84809 hypothetical protein At2g38820 [imported] - Arabidop... 86 3e-16
ref|NP_565896.1| expressed protein; protein id: At2g38820.1, sup... 86 3e-16
ref|NP_188937.1| unknown protein; protein id: At3g22970.1 [Arabi... 78 8e-14
>gb|AAM91628.1| unknown protein [Arabidopsis thaliana]
Length = 341
Score = 86.7 bits (213), Expect = 2e-16
Identities = 53/140 (37%), Positives = 79/140 (55%), Gaps = 23/140 (16%)
Frame = -2
Query: 554 IFVGKCDQLQSIVALVSEAAKQSLRKKGMHVPPWRRVEYVKAKWLSPYSRINEEEKE--- 384
IFVGK D+++ IV++VSEA+KQSL+KKGMH PPWR+ +Y++AKWLS Y+R + E+K
Sbjct: 217 IFVGKSDRIRQIVSIVSEASKQSLKKKGMHFPPWRKADYMRAKWLSSYTRNSGEKKPTVT 276
Query: 383 --------------------KQQVLKEEIENPVAVDVAETVVSGGDGAVAEWKPPELKPK 264
+++VL +++P+ T V D VAE E
Sbjct: 277 SAAKVVAEPELDSSEIELIFEEKVLLPPLKSPI------TSVGRDDDDVAESVKKE---- 326
Query: 263 GSLSGVKVVTGLAVVFDDDN 204
KVVTGLA++F +++
Sbjct: 327 -----AKVVTGLALLFKENH 341
>ref|NP_193198.1| hypothetical protein; protein id: At4g14620.1 [Arabidopsis
thaliana] gi|7485166|pir||G71408 hypothetical protein -
Arabidopsis thaliana gi|2244818|emb|CAB10241.1|
hypothetical protein [Arabidopsis thaliana]
gi|7268168|emb|CAB78504.1| hypothetical protein
[Arabidopsis thaliana]
Length = 358
Score = 86.7 bits (213), Expect = 2e-16
Identities = 53/140 (37%), Positives = 79/140 (55%), Gaps = 23/140 (16%)
Frame = -2
Query: 554 IFVGKCDQLQSIVALVSEAAKQSLRKKGMHVPPWRRVEYVKAKWLSPYSRINEEEKE--- 384
IFVGK D+++ IV++VSEA+KQSL+KKGMH PPWR+ +Y++AKWLS Y+R + E+K
Sbjct: 234 IFVGKSDRIRQIVSIVSEASKQSLKKKGMHFPPWRKADYMRAKWLSSYTRNSGEKKPTVT 293
Query: 383 --------------------KQQVLKEEIENPVAVDVAETVVSGGDGAVAEWKPPELKPK 264
+++VL +++P+ T V D VAE E
Sbjct: 294 SAAKVVAEPELDSSEIELIFEEKVLLPPLKSPI------TSVGRDDDDVAESVKKE---- 343
Query: 263 GSLSGVKVVTGLAVVFDDDN 204
KVVTGLA++F +++
Sbjct: 344 -----AKVVTGLALLFKENH 358
>pir||G84809 hypothetical protein At2g38820 [imported] - Arabidopsis thaliana
Length = 310
Score = 85.9 bits (211), Expect = 3e-16
Identities = 40/75 (53%), Positives = 56/75 (74%), Gaps = 5/75 (6%)
Frame = -2
Query: 554 IFVGKCDQLQSIVALVSEAAKQSLRKKGMHVPPWRRVEYVKAKWLSPYSRIN-----EEE 390
IFVGK D+LQ I+ L+ +AAKQSL+KKG+HVPPWRR EYVK+KWLS + R++ E +
Sbjct: 233 IFVGKADRLQKIIVLICKAAKQSLKKKGLHVPPWRRAEYVKSKWLSSHVRVDQNSNGEVK 292
Query: 389 KEKQQVLKEEIENPV 345
+E +V+ E + + V
Sbjct: 293 QESVEVIAESVSSIV 307
>ref|NP_565896.1| expressed protein; protein id: At2g38820.1, supported by cDNA:
gi_16649128, supported by cDNA: gi_20148590 [Arabidopsis
thaliana] gi|16649129|gb|AAL24416.1| Unknown protein
[Arabidopsis thaliana] gi|20148591|gb|AAM10186.1|
unknown protein [Arabidopsis thaliana]
gi|20198107|gb|AAD25563.2| expressed protein
[Arabidopsis thaliana]
Length = 288
Score = 85.9 bits (211), Expect = 3e-16
Identities = 40/75 (53%), Positives = 56/75 (74%), Gaps = 5/75 (6%)
Frame = -2
Query: 554 IFVGKCDQLQSIVALVSEAAKQSLRKKGMHVPPWRRVEYVKAKWLSPYSRIN-----EEE 390
IFVGK D+LQ I+ L+ +AAKQSL+KKG+HVPPWRR EYVK+KWLS + R++ E +
Sbjct: 211 IFVGKADRLQKIIVLICKAAKQSLKKKGLHVPPWRRAEYVKSKWLSSHVRVDQNSNGEVK 270
Query: 389 KEKQQVLKEEIENPV 345
+E +V+ E + + V
Sbjct: 271 QESVEVIAESVSSIV 285
>ref|NP_188937.1| unknown protein; protein id: At3g22970.1 [Arabidopsis thaliana]
gi|11994729|dbj|BAB03045.1|
gene_id:F5N5.17~pir||G71408~similar to unknown protein
[Arabidopsis thaliana]
Length = 370
Score = 77.8 bits (190), Expect = 8e-14
Identities = 51/139 (36%), Positives = 71/139 (50%), Gaps = 24/139 (17%)
Frame = -2
Query: 554 IFVGKCDQLQSIVALVSEAAKQSLRKKGMHVPPWRRVEYVKAKWLSPYSRIN-------- 399
IFVGK D+L IV L+SEAAKQSL+KKGM PPWR+ EY+++KWLS Y+R +
Sbjct: 242 IFVGKSDRLSQIVFLISEAAKQSLKKKGMPFPPWRKAEYMRSKWLSSYTRASVVVVDETV 301
Query: 398 ---------------EEEKEKQQVLKEEIENP-VAVDVAETVVSGGDGAVAEWKPPELKP 267
+ E + V +E+ +P V V+ + + G D E +
Sbjct: 302 TVTDVTAADAAVEKEVDSVEIELVFEEKCLSPRVIVNSSSSPTDGDDDVAVERE------ 355
Query: 266 KGSLSGVKVVTGLAVVFDD 210
VK VTGLA +F +
Sbjct: 356 ------VKAVTGLASLFKE 368
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 513,657,869
Number of Sequences: 1393205
Number of extensions: 12763043
Number of successful extensions: 124699
Number of sequences better than 10.0: 1931
Number of HSP's better than 10.0 without gapping: 69498
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 103805
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19521267756
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)