Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002887A_C01 KMC002887A_c01
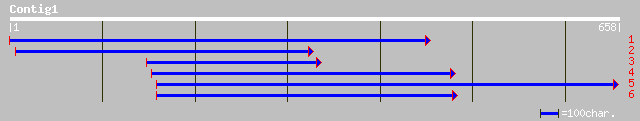
(655 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAK37761.1| zinc transporter protein ZIP1 [Glycine max] 84 2e-15
ref|NP_172022.1| putative zinc transporter; protein id: At1g0530... 67 2e-10
dbj|BAC21508.1| putative zinc transporter protein ZIP1 [Oryza sa... 66 5e-10
ref|NP_187881.1| putative zinc transporter; protein id: At3g1275... 65 8e-10
pir||T52185 zinc transporter ZIP3 [validated] - Arabidopsis thal... 64 2e-09
>gb|AAK37761.1| zinc transporter protein ZIP1 [Glycine max]
Length = 354
Score = 84.0 bits (206), Expect = 2e-15
Identities = 44/52 (84%), Positives = 45/52 (85%)
Frame = -3
Query: 653 FNAASAGIGIYMALADLLAADFMIPRMQNRARLRLGSNLSLLLGAGCMSLLA 498
FNAASAGI IYMAL DLLAADFM PRMQ LRLG+NLSLLLGAGCMSLLA
Sbjct: 300 FNAASAGILIYMALVDLLAADFMNPRMQKSGSLRLGANLSLLLGAGCMSLLA 351
>ref|NP_172022.1| putative zinc transporter; protein id: At1g05300.1 [Arabidopsis
thaliana] gi|25347106|pir||G86187 hypothetical protein
[imported] - Arabidopsis thaliana
gi|2388566|gb|AAB71447.1| Similar to Arabidopsis Fe(II)
transport protein (gb|U27590). [Arabidopsis thaliana]
gi|17385784|gb|AAL38432.1|AF369909_1 putative metal
transporter ZIP5 [Arabidopsis thaliana]
Length = 360
Score = 67.4 bits (163), Expect = 2e-10
Identities = 34/51 (66%), Positives = 43/51 (83%)
Frame = -3
Query: 650 NAASAGIGIYMALADLLAADFMIPRMQNRARLRLGSNLSLLLGAGCMSLLA 498
NAASAGI IYM+L D LAADFM P+MQ+ RL++ +++SLL+GAG MSLLA
Sbjct: 307 NAASAGILIYMSLVDFLAADFMHPKMQSNTRLQIMAHISLLVGAGVMSLLA 357
>dbj|BAC21508.1| putative zinc transporter protein ZIP1 [Oryza sativa (japonica
cultivar-group)] gi|27818099|dbj|BAC55859.1| putative
zinc transporter protein ZIP1 [Oryza sativa (japonica
cultivar-group)]
Length = 357
Score = 65.9 bits (159), Expect = 5e-10
Identities = 32/51 (62%), Positives = 42/51 (81%)
Frame = -3
Query: 650 NAASAGIGIYMALADLLAADFMIPRMQNRARLRLGSNLSLLLGAGCMSLLA 498
N+ +AGI IYMAL DLLA DFM PR+Q++ +L+LG NL++L GAG MS+LA
Sbjct: 304 NSVAAGILIYMALVDLLAEDFMNPRVQSKGKLQLGINLAMLAGAGLMSMLA 354
>ref|NP_187881.1| putative zinc transporter; protein id: At3g12750.1, supported by
cDNA: gi_3252865 [Arabidopsis thaliana]
gi|25347110|pir||T52183 zinc transporter ZIP1
[validated] - Arabidopsis thaliana
gi|3252866|gb|AAC24197.1| putative zinc transporter
[Arabidopsis thaliana] gi|11994417|dbj|BAB02419.1| zinc
transporter-like protein [Arabidopsis thaliana]
Length = 355
Score = 65.1 bits (157), Expect = 8e-10
Identities = 35/51 (68%), Positives = 40/51 (77%)
Frame = -3
Query: 650 NAASAGIGIYMALADLLAADFMIPRMQNRARLRLGSNLSLLLGAGCMSLLA 498
NAASAGI IYM+L DLLA DFM PR+Q+ L L + LSL+LGAG MSLLA
Sbjct: 302 NAASAGILIYMSLVDLLATDFMNPRLQSNLWLHLAAYLSLVLGAGSMSLLA 352
>pir||T52185 zinc transporter ZIP3 [validated] - Arabidopsis thaliana
gi|3252870|gb|AAC24199.1| putative zinc transporter
[Arabidopsis thaliana]
Length = 339
Score = 63.9 bits (154), Expect = 2e-09
Identities = 33/51 (64%), Positives = 42/51 (81%)
Frame = -3
Query: 650 NAASAGIGIYMALADLLAADFMIPRMQNRARLRLGSNLSLLLGAGCMSLLA 498
NAASAGI IYM+L DLLAADF P+MQ+ L++ ++++LLLGAG MSLLA
Sbjct: 286 NAASAGILIYMSLVDLLAADFTHPKMQSNTGLQIMAHIALLLGAGLMSLLA 336
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,375,009
Number of Sequences: 1393205
Number of extensions: 10111344
Number of successful extensions: 27356
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 26313
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27313
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 28144814643
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)