Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
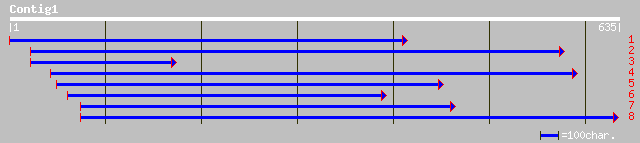
Query= KMC002846A_C01 KMC002846A_c01
(635 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN72008.1| unknown protein [Arabidopsis thaliana] 196 3e-49
ref|NP_173899.1| F-box containing tubby family protein; protein ... 196 3e-49
ref|NP_177816.1| Tub family protein, putative; protein id: At1g7... 196 3e-49
emb|CAB53492.1| CAA303719.1 protein [Oryza sativa] 195 4e-49
dbj|BAC01219.1| putative tubby protein [Oryza sativa (japonica c... 195 5e-49
>gb|AAN72008.1| unknown protein [Arabidopsis thaliana]
Length = 267
Score = 196 bits (497), Expect = 3e-49
Identities = 99/136 (72%), Positives = 110/136 (80%), Gaps = 1/136 (0%)
Frame = -1
Query: 632 STEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTVASVKN 453
S +FSS+RFS++ G ++ EEE RPL+LKNK PRWHEQLQCWCLNFRGRVTVASVKN
Sbjct: 137 SLDFSSSRFSEM-GISCDDNEEEASFRPLILKNKQPRWHEQLQCWCLNFRGRVTVASVKN 195
Query: 452 FQLIAATQPASGGGAPPTGTGAAPPSQPP-SDPDKIILQFGKVGKDMFTMDYRYPLSAFQ 276
FQL+AA QP G TG AAP S P + DK+ILQFGKVGKDMFTMDYRYPLSAFQ
Sbjct: 196 FQLVAARQPQPQG----TGAAAAPTSAPAHPEQDKVILQFGKVGKDMFTMDYRYPLSAFQ 251
Query: 275 AFAICLTSFDTKLACE 228
AFAICL+SFDTKLACE
Sbjct: 252 AFAICLSSFDTKLACE 267
>ref|NP_173899.1| F-box containing tubby family protein; protein id: At1g25280.1
[Arabidopsis thaliana] gi|25518577|pir||E86382
hypothetical protein F4F7.33 [imported] - Arabidopsis
thaliana gi|11067277|gb|AAG28805.1|AC079374_8 unknown
protein [Arabidopsis thaliana]
Length = 445
Score = 196 bits (497), Expect = 3e-49
Identities = 99/136 (72%), Positives = 110/136 (80%), Gaps = 1/136 (0%)
Frame = -1
Query: 632 STEFSSARFSDIIGAGNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTVASVKN 453
S +FSS+RFS++ G ++ EEE RPL+LKNK PRWHEQLQCWCLNFRGRVTVASVKN
Sbjct: 315 SLDFSSSRFSEM-GISCDDNEEEASFRPLILKNKQPRWHEQLQCWCLNFRGRVTVASVKN 373
Query: 452 FQLIAATQPASGGGAPPTGTGAAPPSQPP-SDPDKIILQFGKVGKDMFTMDYRYPLSAFQ 276
FQL+AA QP G TG AAP S P + DK+ILQFGKVGKDMFTMDYRYPLSAFQ
Sbjct: 374 FQLVAARQPQPQG----TGAAAAPTSAPAHPEQDKVILQFGKVGKDMFTMDYRYPLSAFQ 429
Query: 275 AFAICLTSFDTKLACE 228
AFAICL+SFDTKLACE
Sbjct: 430 AFAICLSSFDTKLACE 445
>ref|NP_177816.1| Tub family protein, putative; protein id: At1g76900.1 [Arabidopsis
thaliana] gi|25406477|pir||H96797 hypothetical protein
F22K20.1 [imported] - Arabidopsis thaliana
gi|2829918|gb|AAC00626.1| similar to 'tub' protein
gp|U82468|2072162 [Arabidopsis thaliana]
gi|12322225|gb|AAG51146.1|AC079283_3 Tub family protein,
putative [Arabidopsis thaliana]
gi|22654973|gb|AAM98079.1| At1g76900/F7O12_7
[Arabidopsis thaliana] gi|27764966|gb|AAO23604.1|
At1g76900/F7O12_7 [Arabidopsis thaliana]
Length = 455
Score = 196 bits (497), Expect = 3e-49
Identities = 104/143 (72%), Positives = 111/143 (76%), Gaps = 8/143 (5%)
Frame = -1
Query: 632 STEFSSARFSDIIGAGNEEEE---EEGKER---PLVLKNKSPRWHEQLQCWCLNFRGRVT 471
S +FSSARFSDI+G +E++E EEGKER PLVLKNK PRWHEQLQCWCLNFRGRVT
Sbjct: 313 SNDFSSARFSDILGPLSEDQEVVLEEGKERNSPPLVLKNKPPRWHEQLQCWCLNFRGRVT 372
Query: 470 VASVKNFQLIAATQPASGGGAPPTGTGAAPP--SQPPSDPDKIILQFGKVGKDMFTMDYR 297
VASVKNFQLIAA QP P P S PDKIILQFGKVGKDMFTMD+R
Sbjct: 373 VASVKNFQLIAANQPQPQPQPQPQPQPLTQPQPSGQTDGPDKIILQFGKVGKDMFTMDFR 432
Query: 296 YPLSAFQAFAICLTSFDTKLACE 228
YPLSAFQAFAICL+SFDTKLACE
Sbjct: 433 YPLSAFQAFAICLSSFDTKLACE 455
>emb|CAB53492.1| CAA303719.1 protein [Oryza sativa]
Length = 462
Score = 195 bits (496), Expect = 4e-49
Identities = 105/155 (67%), Positives = 119/155 (76%), Gaps = 19/155 (12%)
Frame = -1
Query: 635 NSTEFSSARFSDIIGAG--NEEEEEEG----KERPLVLKNKSPRWHEQLQCWCLNFRGRV 474
+ST+FSS+RFS+ G +E+E++G KERPLVL+NK+PRWHEQLQCWCLNFRGRV
Sbjct: 308 HSTDFSSSRFSEFGGGALQGQEQEQDGDDVNKERPLVLRNKAPRWHEQLQCWCLNFRGRV 367
Query: 473 TVASVKNFQLIAAT-QPASGGGAPPTGTGAA-----PPSQP-------PSDPDKIILQFG 333
TVASVKNFQLIAA QPASG + P+ G A PSQP S+ D +ILQFG
Sbjct: 368 TVASVKNFQLIAAAPQPASGAASEPSQAGQAAQQQTQPSQPSSSSSSSSSNHDTVILQFG 427
Query: 332 KVGKDMFTMDYRYPLSAFQAFAICLTSFDTKLACE 228
KV KDMFTMDYRYPLSAFQAFAICLTSFDTKLACE
Sbjct: 428 KVAKDMFTMDYRYPLSAFQAFAICLTSFDTKLACE 462
>dbj|BAC01219.1| putative tubby protein [Oryza sativa (japonica cultivar-group)]
Length = 448
Score = 195 bits (495), Expect = 5e-49
Identities = 100/137 (72%), Positives = 112/137 (80%), Gaps = 2/137 (1%)
Frame = -1
Query: 632 STEFSSARFSDIIGA-GNEEEEEEGKERPLVLKNKSPRWHEQLQCWCLNFRGRVTVASVK 456
S +FSSARFSDI G+ ++ E KERPLVL+NK PRWHEQLQCWCLNFRGRVT+ASVK
Sbjct: 320 SMDFSSARFSDISGSIMGGDDNGEIKERPLVLRNKPPRWHEQLQCWCLNFRGRVTIASVK 379
Query: 455 NFQLIAATQPASGGGAPPTGTGAAPPSQP-PSDPDKIILQFGKVGKDMFTMDYRYPLSAF 279
NFQL+AA P PP GA PSQP P+DP+K+ILQFGKV +DMFTMDYRYPLSAF
Sbjct: 380 NFQLVAAPSP------PP--AGAPTPSQPGPADPEKVILQFGKVARDMFTMDYRYPLSAF 431
Query: 278 QAFAICLTSFDTKLACE 228
QAFAICL+SFDTKLACE
Sbjct: 432 QAFAICLSSFDTKLACE 448
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 591,032,880
Number of Sequences: 1393205
Number of extensions: 13957677
Number of successful extensions: 95531
Number of sequences better than 10.0: 129
Number of HSP's better than 10.0 without gapping: 60980
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 86318
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)