Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002844A_C01 KMC002844A_c01
(541 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
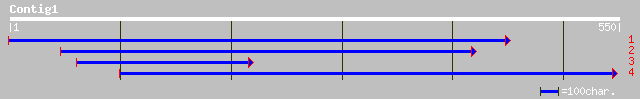
gb|EAA16587.1| R27-2 protein [Plasmodium yoelii yoelii] 35 0.58
gb|EAA21438.1| hypothetical protein [Plasmodium yoelii yoelii] 35 0.58
ref|NP_700715.1| hypothetical protein [Plasmodium falciparum 3D7... 34 1.3
ref|NP_066528.1| ribosomal protein L6 [Naegleria gruberi] gi|104... 33 1.7
ref|NP_008369.1|ND4_13186 NADH dehydrogenase subunit 4 [Onchocer... 33 2.2
>gb|EAA16587.1| R27-2 protein [Plasmodium yoelii yoelii]
Length = 1986
Score = 35.0 bits (79), Expect = 0.58
Identities = 18/48 (37%), Positives = 26/48 (53%)
Frame = +1
Query: 10 YSFIIKKAINRVFTKTSTKDKNKIATKQNSIQKKIQFYGYTSLVLLIH 153
Y +I+ + V TK+ K KNKI QN +QK + F+ L L +H
Sbjct: 356 YEYIVNQIDVIVNTKSQEKLKNKITNYQNDLQKCLMFFNNIFLKLSVH 403
>gb|EAA21438.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 547
Score = 35.0 bits (79), Expect = 0.58
Identities = 33/117 (28%), Positives = 54/117 (45%), Gaps = 2/117 (1%)
Frame = +1
Query: 1 IINYSFIIKKAINRVFTKTSTKDKNKIATKQNSIQKKIQFYGYTSLVLLIHKISLKIHKN 180
I YS+I +K ++R+FT + K I+ K+ QK + Y + + L +N
Sbjct: 301 IQKYSYIFEKELDRIFTPKNIYCKKIISKKEKRAQKGKE---YILEQIKGDDLELNYFEN 357
Query: 181 TLQFHIEILNTTFRILGFVPFHRP*KLDRAKMSFVRLSLSPLNWNKK--KHILGLTI 345
QF I N T+ I FH K DR + + S + LN N ++I+G ++
Sbjct: 358 GYQFFNNITNITYDI-----FHLYNKQDRIFIKTLSSSSNILNINGNVGEYIIGSSL 409
>ref|NP_700715.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23495108|gb|AAN35439.1|AE014833_10 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 1541
Score = 33.9 bits (76), Expect = 1.3
Identities = 10/45 (22%), Positives = 27/45 (59%)
Frame = -1
Query: 187 VMYFYVFLKIFYGLKELNLYNHKIVFFFVLNFV*LLFYFYLLYLF 53
+ +F+ F + + + + +H I+F+F+L + F+F+ +++F
Sbjct: 6 IFFFFFFFDLIFSIASWLILSHYIIFYFILFNLIYSFFFFFIFIF 50
>ref|NP_066528.1| ribosomal protein L6 [Naegleria gruberi]
gi|10444240|gb|AAG17806.1|AF288092_31 ribosomal protein
L6 [Naegleria gruberi]
Length = 166
Score = 33.5 bits (75), Expect = 1.7
Identities = 18/55 (32%), Positives = 30/55 (53%), Gaps = 3/55 (5%)
Frame = -1
Query: 217 MLYLKFQYETVMYFYVFLKIFYGLKELNLYNHKI---VFFFVLNFV*LLFYFYLL 62
M + +F Y +YFYV L + +L+ + +K FFF+++ + FYF LL
Sbjct: 1 MSFKQFFYINNLYFYVELNTYLVGTDLSFFKYKFNSSFFFFIMDNIFYFFYFNLL 55
>ref|NP_008369.1|ND4_13186 NADH dehydrogenase subunit 4 [Onchocerca volvulus]
gi|7432353|pir||T11064 NADH2 dehydrogenase (ubiquinone)
(EC 1.6.5.3) chain 4 - nematode (Onchocerca volvulus)
mitochondrion gi|2735936|gb|AAC61611.1| NADH
dehydrogenase subunit 4 [Onchocerca volvulus]
Length = 410
Score = 33.1 bits (74), Expect = 2.2
Identities = 19/43 (44%), Positives = 28/43 (64%)
Frame = -1
Query: 181 YFYVFLKIFYGLKELNLYNHKIVFFFVLNFV*LLFYFYLLYLF 53
Y+ +F +F+G+ L LY+H VFFF LNFV + F++ Y F
Sbjct: 123 YYLIFYTLFFGMPYLFLYSH--VFFF-LNFV--YYDFFVSYEF 160
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 441,811,825
Number of Sequences: 1393205
Number of extensions: 9286470
Number of successful extensions: 22604
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 21488
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22490
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)