Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002832A_C02 KMC002832A_c02
(770 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
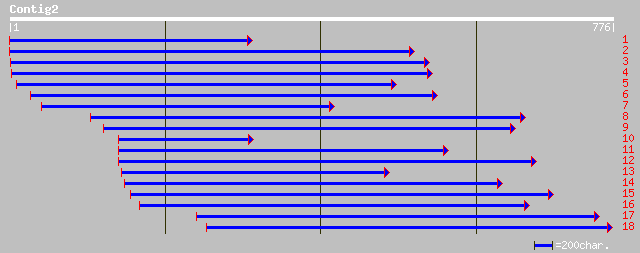
emb|CAD80089.1| hypothetical protein [Momordica charantia] 87 3e-16
dbj|BAB56058.1| P0665A11.3 [Oryza sativa (japonica cultivar-grou... 73 4e-12
gb|AAM61209.1| unknown [Arabidopsis thaliana] 56 7e-07
ref|NP_565199.1| expressed protein; protein id: At1g79090.1 [Ara... 56 7e-07
pir||T01046 hypothetical protein YUP8H12R.29 - Arabidopsis thali... 56 7e-07
>emb|CAD80089.1| hypothetical protein [Momordica charantia]
Length = 90
Score = 86.7 bits (213), Expect = 3e-16
Identities = 45/75 (60%), Positives = 60/75 (80%)
Frame = -2
Query: 769 SIMQSLVTQGAPNVAVIGSDAAKAISKEMPVELLRASLPHTDDHPRKLLLDFAQRSVPVV 590
+I+QSL +Q + +I S+AA+AI +EMPVELLRASLPHT++ RKLL+DFAQRS+PV
Sbjct: 14 TIVQSLFSQTPSSTDIIESEAARAIGREMPVELLRASLPHTNEPQRKLLMDFAQRSMPVS 73
Query: 589 GLSSYAGSSSGHVNS 545
G S++ G SSG +NS
Sbjct: 74 GFSAH-GGSSGQMNS 87
>dbj|BAB56058.1| P0665A11.3 [Oryza sativa (japonica cultivar-group)]
gi|21743313|dbj|BAC03308.1| B1143G03.11 [Oryza sativa
(japonica cultivar-group)]
Length = 763
Score = 73.2 bits (178), Expect = 4e-12
Identities = 38/63 (60%), Positives = 48/63 (75%)
Frame = -2
Query: 769 SIMQSLVTQGAPNVAVIGSDAAKAISKEMPVELLRASLPHTDDHPRKLLLDFAQRSVPVV 590
SI Q V Q +V GS+ +KA S+EMPVELLRASLPHT+D R+LLLDFAQR++PV
Sbjct: 663 SIKQMFVMQSP--CSVTGSEVSKATSREMPVELLRASLPHTNDQQRQLLLDFAQRTMPVT 720
Query: 589 GLS 581
G++
Sbjct: 721 GIN 723
>gb|AAM61209.1| unknown [Arabidopsis thaliana]
Length = 148
Score = 55.8 bits (133), Expect = 7e-07
Identities = 30/58 (51%), Positives = 43/58 (73%), Gaps = 1/58 (1%)
Frame = -2
Query: 769 SIMQSLVTQGAPNVAV-IGSDAAKAISKEMPVELLRASLPHTDDHPRKLLLDFAQRSV 599
SIMQSL Q P+ A I +AAKAI +EMP+ELLR+S PH D+ +++L++F +RS+
Sbjct: 82 SIMQSL--QLPPHFATEISEEAAKAIVREMPIELLRSSFPHIDEQQKRILMEFLKRSM 137
>ref|NP_565199.1| expressed protein; protein id: At1g79090.1 [Arabidopsis thaliana]
Length = 793
Score = 55.8 bits (133), Expect = 7e-07
Identities = 30/58 (51%), Positives = 43/58 (73%), Gaps = 1/58 (1%)
Frame = -2
Query: 769 SIMQSLVTQGAPNVAV-IGSDAAKAISKEMPVELLRASLPHTDDHPRKLLLDFAQRSV 599
SIMQSL Q P+ A I +AAKAI +EMP+ELLR+S PH D+ +++L++F +RS+
Sbjct: 727 SIMQSL--QLPPHFATEISEEAAKAIVREMPIELLRSSFPHIDEQQKRILMEFLKRSM 782
>pir||T01046 hypothetical protein YUP8H12R.29 - Arabidopsis thaliana
gi|3152568|gb|AAC17049.1| Similar to hypothetical
protein product gb|Z97337 from A. thaliana. EST
gb|H76597 comes from this gene. [Arabidopsis thaliana]
Length = 841
Score = 55.8 bits (133), Expect = 7e-07
Identities = 30/58 (51%), Positives = 43/58 (73%), Gaps = 1/58 (1%)
Frame = -2
Query: 769 SIMQSLVTQGAPNVAV-IGSDAAKAISKEMPVELLRASLPHTDDHPRKLLLDFAQRSV 599
SIMQSL Q P+ A I +AAKAI +EMP+ELLR+S PH D+ +++L++F +RS+
Sbjct: 775 SIMQSL--QLPPHFATEISEEAAKAIVREMPIELLRSSFPHIDEQQKRILMEFLKRSM 830
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 649,075,909
Number of Sequences: 1393205
Number of extensions: 14199916
Number of successful extensions: 29510
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 28573
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29506
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 37815044670
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)