Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002820A_C01 KMC002820A_c01
(563 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
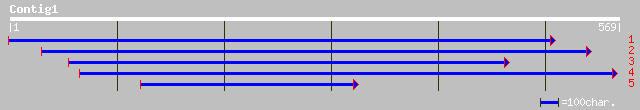
ref|NP_197131.2| putative protein; protein id: At5g16270.1, supp... 84 9e-16
gb|AAK96855.1| putative protein [Arabidopsis thaliana] gi|183774... 84 9e-16
pir||T51497 hypothetical protein T21H19_190 - Arabidopsis thalia... 84 9e-16
gb|AAG44843.1|AF281155_1 cohesion family protein SYN3 [Arabidops... 59 4e-08
ref|NP_191514.1| putative protein; protein id: At3g59550.1, supp... 59 4e-08
>ref|NP_197131.2| putative protein; protein id: At5g16270.1, supported by cDNA:
gi_18157648 [Arabidopsis thaliana]
gi|18157649|gb|AAL62060.1|AF400129_1 RAD21-3 [Arabidopsis
thaliana]
Length = 1031
Score = 84.3 bits (207), Expect = 9e-16
Identities = 40/55 (72%), Positives = 47/55 (84%)
Frame = -1
Query: 554 LKLDNILAGKTRKEASRMFFETLVLKTRDYVHVEQAEPYDNIDLKPRPKLMKADF 390
L D +LAGKTRKEASRMFFETLVLKTRDY+ VEQ +PY++I +KPRPKL K+ F
Sbjct: 977 LVADKLLAGKTRKEASRMFFETLVLKTRDYIQVEQGKPYESIIIKPRPKLTKSIF 1031
>gb|AAK96855.1| putative protein [Arabidopsis thaliana] gi|18377436|gb|AAL66884.1|
putative protein [Arabidopsis thaliana]
Length = 236
Score = 84.3 bits (207), Expect = 9e-16
Identities = 40/55 (72%), Positives = 47/55 (84%)
Frame = -1
Query: 554 LKLDNILAGKTRKEASRMFFETLVLKTRDYVHVEQAEPYDNIDLKPRPKLMKADF 390
L D +LAGKTRKEASRMFFETLVLKTRDY+ VEQ +PY++I +KPRPKL K+ F
Sbjct: 182 LVADKLLAGKTRKEASRMFFETLVLKTRDYIQVEQGKPYESIIIKPRPKLTKSVF 236
>pir||T51497 hypothetical protein T21H19_190 - Arabidopsis thaliana
gi|9755837|emb|CAC01868.1| putative protein [Arabidopsis
thaliana]
Length = 1021
Score = 84.3 bits (207), Expect = 9e-16
Identities = 40/55 (72%), Positives = 47/55 (84%)
Frame = -1
Query: 554 LKLDNILAGKTRKEASRMFFETLVLKTRDYVHVEQAEPYDNIDLKPRPKLMKADF 390
L D +LAGKTRKEASRMFFETLVLKTRDY+ VEQ +PY++I +KPRPKL K+ F
Sbjct: 967 LVADKLLAGKTRKEASRMFFETLVLKTRDYIQVEQGKPYESIIIKPRPKLTKSIF 1021
>gb|AAG44843.1|AF281155_1 cohesion family protein SYN3 [Arabidopsis thaliana]
Length = 692
Score = 58.9 bits (141), Expect = 4e-08
Identities = 30/51 (58%), Positives = 37/51 (71%)
Frame = -1
Query: 557 NLKLDNILAGKTRKEASRMFFETLVLKTRDYVHVEQAEPYDNIDLKPRPKL 405
+L L ILAGKTRK A+RMFFETLVLK+R + ++Q PY +I LK P L
Sbjct: 636 DLSLSEILAGKTRKLAARMFFETLVLKSRGLIDMQQDRPYGDIALKLMPAL 686
>ref|NP_191514.1| putative protein; protein id: At3g59550.1, supported by cDNA:
gi_12006361, supported by cDNA: gi_18157646 [Arabidopsis
thaliana] gi|11358049|pir||T49296 hypothetical protein
T16L24.100 - Arabidopsis thaliana
gi|6996291|emb|CAB75452.1| putative protein [Arabidopsis
thaliana] gi|18157647|gb|AAL62059.1|AF400128_1 RAD21-2
[Arabidopsis thaliana]
Length = 693
Score = 58.9 bits (141), Expect = 4e-08
Identities = 30/51 (58%), Positives = 37/51 (71%)
Frame = -1
Query: 557 NLKLDNILAGKTRKEASRMFFETLVLKTRDYVHVEQAEPYDNIDLKPRPKL 405
+L L ILAGKTRK A+RMFFETLVLK+R + ++Q PY +I LK P L
Sbjct: 637 DLSLSEILAGKTRKLAARMFFETLVLKSRGLIDMQQDRPYGDIALKLMPAL 687
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 507,005,358
Number of Sequences: 1393205
Number of extensions: 11329381
Number of successful extensions: 22718
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 22081
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22707
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)