Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002808A_C01 KMC002808A_c01
(823 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
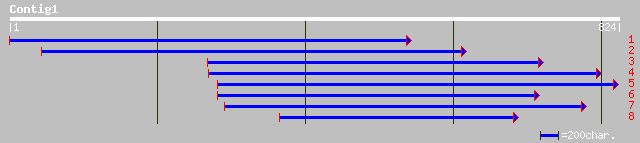
dbj|BAA78744.1| EST AU065533(C2174) corresponds to a region of t... 222 6e-57
pir||B96832 hypothetical protein F18B13.15 [imported] - Arabidop... 212 6e-54
ref|NP_178124.1| splicing factor Prp8, putative; protein id: At1... 212 6e-54
ref|NP_195589.1| splicing factor - like protein; protein id: At4... 200 2e-50
gb|EAA04255.1| ebiP5721 [Anopheles gambiae str. PEST] 178 1e-43
>dbj|BAA78744.1| EST AU065533(C2174) corresponds to a region of the predicted
gene.~Similar to Homo sapiens splicing factor Prp8 mRNA,
complete cds.(AF092565) [Oryza sativa (japonica
cultivar-group)]
Length = 2391
Score = 222 bits (565), Expect = 6e-57
Identities = 101/119 (84%), Positives = 111/119 (92%)
Frame = -1
Query: 823 TPGSCSLTAYKLTPSGYEWGRINKDTGSNPHGYLPTHYEKVQMLLSDRFLGFYMIPDNGP 644
TPGSCSLTAYKLTPSGYEWGR NKDTGSNPHGYLPTHYEKVQMLLSDRFLGFYM+PDN P
Sbjct: 2245 TPGSCSLTAYKLTPSGYEWGRSNKDTGSNPHGYLPTHYEKVQMLLSDRFLGFYMVPDNTP 2304
Query: 643 WNYNFMGVRHAPSMKYGVKLGTPREYYNEDHRPTHFLEFSNMEEKGETVVEGDREDTFS 467
WN+NFMGV+H P MKY +KLGTPR++Y+EDHRPTHFLEFSN++E GE V EGDREDTF+
Sbjct: 2305 WNFNFMGVKHDPLMKYNMKLGTPRDFYHEDHRPTHFLEFSNIDE-GE-VAEGDREDTFT 2361
>pir||B96832 hypothetical protein F18B13.15 [imported] - Arabidopsis thaliana
gi|5902365|gb|AAD55467.1|AC009322_7 Putative splicing
factor Prp8 [Arabidopsis thaliana]
Length = 2359
Score = 212 bits (539), Expect = 6e-54
Identities = 93/119 (78%), Positives = 108/119 (90%)
Frame = -1
Query: 823 TPGSCSLTAYKLTPSGYEWGRINKDTGSNPHGYLPTHYEKVQMLLSDRFLGFYMIPDNGP 644
TPGSCSLT+YKLT +GYEWGR+NKD GSNPHGYLPTHYEKVQMLLSDRFLGFYM+P++GP
Sbjct: 2243 TPGSCSLTSYKLTQTGYEWGRLNKDNGSNPHGYLPTHYEKVQMLLSDRFLGFYMVPESGP 2302
Query: 643 WNYNFMGVRHAPSMKYGVKLGTPREYYNEDHRPTHFLEFSNMEEKGETVVEGDREDTFS 467
WNY+F GV+H SMKY VKLG+P+E+Y+E+HRPTHFLEFSNMEE + EGDREDTF+
Sbjct: 2303 WNYSFTGVKHTLSMKYSVKLGSPKEFYHEEHRPTHFLEFSNMEE--ADITEGDREDTFT 2359
>ref|NP_178124.1| splicing factor Prp8, putative; protein id: At1g80070.1 [Arabidopsis
thaliana]
Length = 2382
Score = 212 bits (539), Expect = 6e-54
Identities = 93/119 (78%), Positives = 108/119 (90%)
Frame = -1
Query: 823 TPGSCSLTAYKLTPSGYEWGRINKDTGSNPHGYLPTHYEKVQMLLSDRFLGFYMIPDNGP 644
TPGSCSLT+YKLT +GYEWGR+NKD GSNPHGYLPTHYEKVQMLLSDRFLGFYM+P++GP
Sbjct: 2266 TPGSCSLTSYKLTQTGYEWGRLNKDNGSNPHGYLPTHYEKVQMLLSDRFLGFYMVPESGP 2325
Query: 643 WNYNFMGVRHAPSMKYGVKLGTPREYYNEDHRPTHFLEFSNMEEKGETVVEGDREDTFS 467
WNY+F GV+H SMKY VKLG+P+E+Y+E+HRPTHFLEFSNMEE + EGDREDTF+
Sbjct: 2326 WNYSFTGVKHTLSMKYSVKLGSPKEFYHEEHRPTHFLEFSNMEE--ADITEGDREDTFT 2382
>ref|NP_195589.1| splicing factor - like protein; protein id: At4g38780.1 [Arabidopsis
thaliana] gi|7488347|pir||T06077 splicing factor PRP8
homolog T9A14.60 - Arabidopsis thaliana
gi|4490330|emb|CAB38612.1| splicing factor-like protein
[Arabidopsis thaliana] gi|7270861|emb|CAB80541.1|
splicing factor-like protein [Arabidopsis thaliana]
Length = 2352
Score = 200 bits (509), Expect = 2e-50
Identities = 87/119 (73%), Positives = 102/119 (85%)
Frame = -1
Query: 823 TPGSCSLTAYKLTPSGYEWGRINKDTGSNPHGYLPTHYEKVQMLLSDRFLGFYMIPDNGP 644
TPGSCSLT+YKLT +GYEWGR+NKDTGSNPHGYLPTHYEKVQMLLSDRF GFYM+P+NGP
Sbjct: 2238 TPGSCSLTSYKLTQAGYEWGRLNKDTGSNPHGYLPTHYEKVQMLLSDRFFGFYMVPENGP 2297
Query: 643 WNYNFMGVRHAPSMKYGVKLGTPREYYNEDHRPTHFLEFSNMEEKGETVVEGDREDTFS 467
WNYNFMG H S+ Y + LGTP+EYY++ HRPTHFL+FS MEE G+ DR+D+F+
Sbjct: 2298 WNYNFMGANHTVSINYSLTLGTPKEYYHQVHRPTHFLQFSKMEEDGDL----DRDDSFA 2352
>gb|EAA04255.1| ebiP5721 [Anopheles gambiae str. PEST]
Length = 2387
Score = 178 bits (451), Expect = 1e-43
Identities = 80/118 (67%), Positives = 97/118 (81%)
Frame = -1
Query: 823 TPGSCSLTAYKLTPSGYEWGRINKDTGSNPHGYLPTHYEKVQMLLSDRFLGFYMIPDNGP 644
TPGSCSLTAYKLTPSGYEWG N D G+NP GYLP+HYE+VQMLLS++FLGF+M+P G
Sbjct: 2271 TPGSCSLTAYKLTPSGYEWGHKNTDKGNNPKGYLPSHYERVQMLLSNKFLGFFMVPAQGS 2330
Query: 643 WNYNFMGVRHAPSMKYGVKLGTPREYYNEDHRPTHFLEFSNMEEKGETVVEGDREDTF 470
WNYNFMGVRH P+MKY ++L P+E+Y+E HRP+HFL FS++EE G+ DRED F
Sbjct: 2331 WNYNFMGVRHDPNMKYELQLANPKEFYHEIHRPSHFLLFSSLEEGGDG-NGADREDMF 2387
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 735,666,377
Number of Sequences: 1393205
Number of extensions: 16396703
Number of successful extensions: 34675
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 33476
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34659
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42576939184
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)