Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002796A_C01 KMC002796A_c01
(562 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
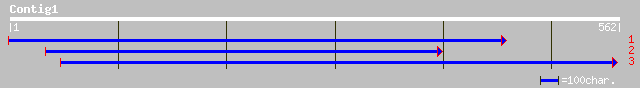
dbj|BAC41893.1| putative importin beta [Arabidopsis thaliana] 216 2e-55
ref|NP_200160.1| importin beta; protein id: At5g53480.1 [Arabido... 216 2e-55
dbj|BAA34862.1| importin-beta2 [Oryza sativa (japonica cultivar-... 207 5e-53
dbj|BAA34861.1| importin-beta1 [Oryza sativa (japonica cultivar-... 165 3e-40
emb|CAC79691.1| Importin beta-like protein [Oryza sativa (indica... 155 2e-37
>dbj|BAC41893.1| putative importin beta [Arabidopsis thaliana]
Length = 808
Score = 216 bits (550), Expect = 2e-55
Identities = 108/142 (76%), Positives = 126/142 (88%)
Frame = +2
Query: 137 MAMEVTQILLNAQAVDGAVRKQAEDNLKQFQEQNLPSFLYSLAGELANDDKPAESRKLAG 316
MAMEVTQ+L+NAQ++DG VRK AE++LKQFQEQNL FL SLAGELAND+KP +SRKLAG
Sbjct: 1 MAMEVTQLLINAQSIDGTVRKHAEESLKQFQEQNLAGFLLSLAGELANDEKPVDSRKLAG 60
Query: 317 LILKNALDAKEQHRKVELIQRWLSLDPALKAQIKAFLLRTLSSLSLDARSTASQVIAKVA 496
L+LKNALDAKEQHRK EL+QRWL+LD + K+QI+AFLL+TLS+ D RSTASQVIAKVA
Sbjct: 61 LVLKNALDAKEQHRKYELVQRWLALDMSTKSQIRAFLLKTLSAPVPDVRSTASQVIAKVA 120
Query: 497 GIELPHKEWPEWVGSLLSNIHQ 562
GIELP K+WPE + SLLSNIHQ
Sbjct: 121 GIELPQKQWPELIVSLLSNIHQ 142
>ref|NP_200160.1| importin beta; protein id: At5g53480.1 [Arabidopsis thaliana]
gi|9759187|dbj|BAB09724.1| importin beta [Arabidopsis
thaliana]
Length = 870
Score = 216 bits (550), Expect = 2e-55
Identities = 108/142 (76%), Positives = 126/142 (88%)
Frame = +2
Query: 137 MAMEVTQILLNAQAVDGAVRKQAEDNLKQFQEQNLPSFLYSLAGELANDDKPAESRKLAG 316
MAMEVTQ+L+NAQ++DG VRK AE++LKQFQEQNL FL SLAGELAND+KP +SRKLAG
Sbjct: 1 MAMEVTQLLINAQSIDGTVRKHAEESLKQFQEQNLAGFLLSLAGELANDEKPVDSRKLAG 60
Query: 317 LILKNALDAKEQHRKVELIQRWLSLDPALKAQIKAFLLRTLSSLSLDARSTASQVIAKVA 496
L+LKNALDAKEQHRK EL+QRWL+LD + K+QI+AFLL+TLS+ D RSTASQVIAKVA
Sbjct: 61 LVLKNALDAKEQHRKYELVQRWLALDMSTKSQIRAFLLKTLSAPVPDVRSTASQVIAKVA 120
Query: 497 GIELPHKEWPEWVGSLLSNIHQ 562
GIELP K+WPE + SLLSNIHQ
Sbjct: 121 GIELPQKQWPELIVSLLSNIHQ 142
>dbj|BAA34862.1| importin-beta2 [Oryza sativa (japonica cultivar-group)]
Length = 872
Score = 207 bits (528), Expect = 5e-53
Identities = 103/142 (72%), Positives = 124/142 (86%)
Frame = +2
Query: 137 MAMEVTQILLNAQAVDGAVRKQAEDNLKQFQEQNLPSFLYSLAGELANDDKPAESRKLAG 316
M++++TQ+LL+AQ+ DGA RK AE++LKQFQEQNLP FL+SL+ ELAN++KP ESR+LAG
Sbjct: 1 MSLDITQVLLSAQSPDGATRKLAEESLKQFQEQNLPGFLFSLSNELANEEKPEESRRLAG 60
Query: 317 LILKNALDAKEQHRKVELIQRWLSLDPALKAQIKAFLLRTLSSLSLDARSTASQVIAKVA 496
LILKNALDAKEQHRK EL QRWL+LD +KAQIK FLL+TLSS ARST+SQVIAKVA
Sbjct: 61 LILKNALDAKEQHRKNELFQRWLALDVGVKAQIKGFLLQTLSSPVASARSTSSQVIAKVA 120
Query: 497 GIELPHKEWPEWVGSLLSNIHQ 562
GIE+P K+WPE + SLLSNIHQ
Sbjct: 121 GIEIPQKQWPELIASLLSNIHQ 142
>dbj|BAA34861.1| importin-beta1 [Oryza sativa (japonica cultivar-group)]
Length = 868
Score = 165 bits (418), Expect = 3e-40
Identities = 85/140 (60%), Positives = 110/140 (77%)
Frame = +2
Query: 143 MEVTQILLNAQAVDGAVRKQAEDNLKQFQEQNLPSFLYSLAGELANDDKPAESRKLAGLI 322
M +TQILL+AQ+ DG +R AE NLKQFQEQNLP+FL SL+ EL++++KP ESR+LAG+I
Sbjct: 1 MNITQILLSAQSADGNLRVVAEGNLKQFQEQNLPNFLLSLSVELSDNEKPPESRRLAGII 60
Query: 323 LKNALDAKEQHRKVELIQRWLSLDPALKAQIKAFLLRTLSSLSLDARSTASQVIAKVAGI 502
LKN+LDAK+ +K LIQ+W+SLDP++K +IK LL TL S DAR T+SQVIAKVA I
Sbjct: 61 LKNSLDAKDSAKKELLIQQWVSLDPSIKQKIKESLLITLGSSVHDARHTSSQVIAKVASI 120
Query: 503 ELPHKEWPEWVGSLLSNIHQ 562
E+P +EW E + LL N+ Q
Sbjct: 121 EIPRREWQELIAKLLGNMTQ 140
>emb|CAC79691.1| Importin beta-like protein [Oryza sativa (indica cultivar-group)]
Length = 864
Score = 155 bits (393), Expect = 2e-37
Identities = 80/134 (59%), Positives = 105/134 (77%)
Frame = +2
Query: 161 LLNAQAVDGAVRKQAEDNLKQFQEQNLPSFLYSLAGELANDDKPAESRKLAGLILKNALD 340
LL+AQ+ DG +R AE NLKQFQEQNLP+FL SL+ EL++++KP ESR+LAG+ILKN+LD
Sbjct: 3 LLSAQSADGNLRVVAEGNLKQFQEQNLPNFLLSLSVELSDNEKPPESRRLAGIILKNSLD 62
Query: 341 AKEQHRKVELIQRWLSLDPALKAQIKAFLLRTLSSLSLDARSTASQVIAKVAGIELPHKE 520
AK+ +K LIQ+W+SLDP++K +IK LL TL S DAR T+SQVIAKVA IE+P +E
Sbjct: 63 AKDSAKKELLIQQWVSLDPSIKQKIKESLLITLGSSVHDARHTSSQVIAKVASIEIPRRE 122
Query: 521 WPEWVGSLLSNIHQ 562
W + + LL N+ Q
Sbjct: 123 WQQLIAKLLGNMTQ 136
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,046,355
Number of Sequences: 1393205
Number of extensions: 10131180
Number of successful extensions: 31476
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 30550
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31450
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)