Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002792A_C01 KMC002792A_c01
(576 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
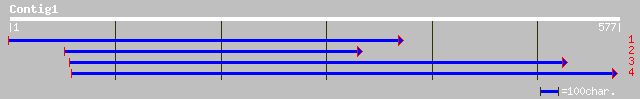
ref|NP_199849.1| putative protein; protein id: At5g50380.1 [Arab... 137 7e-32
dbj|BAB86177.1| putative leucine zipper protein [Oryza sativa (j... 107 1e-22
ref|NP_177391.1| unknown protein; protein id: At1g72470.1 [Arabi... 104 7e-22
ref|NP_566477.1| Expressed protein; protein id: At3g14090.1, sup... 101 8e-21
gb|AAL07238.2| unknown protein [Arabidopsis thaliana] 101 8e-21
>ref|NP_199849.1| putative protein; protein id: At5g50380.1 [Arabidopsis thaliana]
gi|9758920|dbj|BAB09457.1|
gb|AAD25781.1~gene_id:MXI22.10~strong similarity to
unknown protein [Arabidopsis thaliana]
Length = 637
Score = 137 bits (346), Expect = 7e-32
Identities = 67/86 (77%), Positives = 77/86 (88%), Gaps = 1/86 (1%)
Frame = -1
Query: 576 ACFEEIYRVQTAWKVPDEQLREEMRISISEKVIPAYRSFVGRFKSQLE-GRHSGKYIKYT 400
A FEE+YR+QTAWKVPD QLREE+RISISEKVIPAYR+F GR +SQLE GRH+GKYIKYT
Sbjct: 552 ASFEELYRLQTAWKVPDPQLREELRISISEKVIPAYRAFFGRNRSQLEGGRHAGKYIKYT 611
Query: 399 PEDLETYLLDLFEGSPAVLHHIRRKS 322
P+DLE+YL DLFEG+ V+HH RRKS
Sbjct: 612 PDDLESYLPDLFEGNQLVIHHPRRKS 637
>dbj|BAB86177.1| putative leucine zipper protein [Oryza sativa (japonica
cultivar-group)]
Length = 646
Score = 107 bits (267), Expect = 1e-22
Identities = 51/72 (70%), Positives = 60/72 (82%), Gaps = 1/72 (1%)
Frame = -1
Query: 570 FEEIYRVQTAWKVPDEQLREEMRISISEKVIPAYRSFVGRFKSQLE-GRHSGKYIKYTPE 394
FEE+Y+ QT WKV D QLREE++ISISEKV+PAYRSFVGRF+ QLE GR+S +YIKY PE
Sbjct: 570 FEELYKTQTTWKVVDPQLREELKISISEKVLPAYRSFVGRFRGQLEGGRNSARYIKYNPE 629
Query: 393 DLETYLLDLFEG 358
DLE + D FEG
Sbjct: 630 DLENQVSDFFEG 641
>ref|NP_177391.1| unknown protein; protein id: At1g72470.1 [Arabidopsis thaliana]
gi|25308572|pir||H96748 unknown protein T10D10.6
[imported] - Arabidopsis thaliana
gi|12325284|gb|AAG52591.1|AC016529_22 unknown protein;
29470-27569 [Arabidopsis thaliana]
Length = 633
Score = 104 bits (260), Expect = 7e-22
Identities = 53/86 (61%), Positives = 67/86 (77%), Gaps = 1/86 (1%)
Frame = -1
Query: 576 ACFEEIYRVQTAWKVPDEQLREEMRISISEKVIPAYRSFVGRFKSQLE-GRHSGKYIKYT 400
A FEE++RVQ+ W VPD QLREE++ISI EK+ PAYRSF+GRF+S +E G+H YIK +
Sbjct: 548 ALFEEVHRVQSQWLVPDSQLREELKISILEKLSPAYRSFLGRFRSHIESGKHPENYIKIS 607
Query: 399 PEDLETYLLDLFEGSPAVLHHIRRKS 322
EDLET +LDLFEG A H+RR+S
Sbjct: 608 VEDLETEVLDLFEGYSAT-QHLRRRS 632
>ref|NP_566477.1| Expressed protein; protein id: At3g14090.1, supported by cDNA:
gi_15810240 [Arabidopsis thaliana]
Length = 451
Score = 101 bits (251), Expect = 8e-21
Identities = 48/82 (58%), Positives = 61/82 (73%), Gaps = 1/82 (1%)
Frame = -1
Query: 570 FEEIYRVQTAWKVPDEQLREEMRISISEKVIPAYRSFVGRFKSQLE-GRHSGKYIKYTPE 394
FEE++R Q+ W VPD QLREE+RIS+SE +IPAYRSF+GRF+ +E GRH Y+KY+ E
Sbjct: 370 FEEVHRTQSTWSVPDAQLREELRISLSEHLIPAYRSFLGRFRGHIESGRHPENYLKYSVE 429
Query: 393 DLETYLLDLFEGSPAVLHHIRR 328
D+ET +LD FEG H RR
Sbjct: 430 DIETIVLDFFEGYTTPPHLRRR 451
>gb|AAL07238.2| unknown protein [Arabidopsis thaliana]
Length = 503
Score = 101 bits (251), Expect = 8e-21
Identities = 48/82 (58%), Positives = 61/82 (73%), Gaps = 1/82 (1%)
Frame = -1
Query: 570 FEEIYRVQTAWKVPDEQLREEMRISISEKVIPAYRSFVGRFKSQLE-GRHSGKYIKYTPE 394
FEE++R Q+ W VPD QLREE+RIS+SE +IPAYRSF+GRF+ +E GRH Y+KY+ E
Sbjct: 422 FEEVHRTQSTWSVPDAQLREELRISLSEHLIPAYRSFLGRFRGHIESGRHPENYLKYSVE 481
Query: 393 DLETYLLDLFEGSPAVLHHIRR 328
D+ET +LD FEG H RR
Sbjct: 482 DIETIVLDFFEGYTTPPHLRRR 503
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 497,517,559
Number of Sequences: 1393205
Number of extensions: 11013905
Number of successful extensions: 23060
Number of sequences better than 10.0: 65
Number of HSP's better than 10.0 without gapping: 22295
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23024
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)