Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002773A_C01 KMC002773A_c01
(560 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
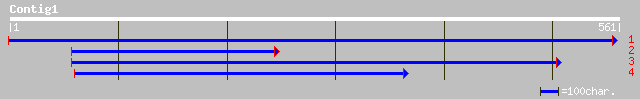
ref|NP_186827.1| unknown protein; protein id: At3g01780.1 [Arabi... 97 1e-19
gb|AAO42242.1| unknown protein [Arabidopsis thaliana] 50 1e-05
ref|NP_036881.1| nucleolin [Rattus norvegicus] gi|128844|sp|P133... 43 0.002
pir||JH0148 nucleolin - rat 43 0.002
dbj|BAC34476.1| unnamed protein product [Mus musculus] 42 0.004
>ref|NP_186827.1| unknown protein; protein id: At3g01780.1 [Arabidopsis thaliana]
gi|6016734|gb|AAF01560.1|AC009325_30 unknown protein
[Arabidopsis thaliana]
gi|6091721|gb|AAF03433.1|AC010797_9 unknown protein
[Arabidopsis thaliana]
Length = 1192
Score = 97.1 bits (240), Expect = 1e-19
Identities = 58/105 (55%), Positives = 71/105 (67%), Gaps = 19/105 (18%)
Frame = -2
Query: 559 ERLRISMERIALLKPAQPRPKTPK--------------SDDEDEEEDKDKEKKDGEEDEK 422
E+L+ISMERIALLK AQP+ KT K +D+DEE + KEK++G++ E+
Sbjct: 1089 EKLKISMERIALLKAAQPK-KTSKIEEESENEEEEEGEEEDDDEEVKEKKEKEEGKDKEE 1147
Query: 421 KKGPS----TLSKLTAEEAEHQALQAAVLQEWHMLCKDRS-TEVN 302
KK T SKLTAEE EH ALQAAVLQEWH+LCKDR T+VN
Sbjct: 1148 KKKKEKEKGTFSKLTAEETEHMALQAAVLQEWHILCKDRKYTKVN 1192
>gb|AAO42242.1| unknown protein [Arabidopsis thaliana]
Length = 1135
Score = 50.4 bits (119), Expect = 1e-05
Identities = 27/49 (55%), Positives = 40/49 (81%), Gaps = 1/49 (2%)
Frame = -2
Query: 559 ERLRISMERIALLKPAQPRPKTPKSDDEDE-EEDKDKEKKDGEEDEKKK 416
E+L+ISMERIALLK AQP+ KT K ++E E EE+++ E++D +E+ K+K
Sbjct: 1073 EKLKISMERIALLKAAQPK-KTSKIEEESENEEEEEGEEEDDDEEVKEK 1120
>ref|NP_036881.1| nucleolin [Rattus norvegicus] gi|128844|sp|P13383|NUCL_RAT
Nucleolin (Protein C23) gi|205792|gb|AAA41732.1|
nucleolin
Length = 713
Score = 43.1 bits (100), Expect = 0.002
Identities = 20/59 (33%), Positives = 35/59 (58%)
Frame = -2
Query: 523 LKPAQPRPKTPKSDDEDEEEDKDKEKKDGEEDEKKKGPSTLSKLTAEEAEHQALQAAVL 347
+KPA+ P P S+DEDEE+D D++ D +E+E+++ S + A+ + A V+
Sbjct: 175 VKPAKAAPAAPASEDEDEEDDDDEDDDDDDEEEEEEDDSEEEVMEITPAKGKKTPAKVV 233
Score = 34.3 bits (77), Expect = 1.1
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Frame = -2
Query: 520 KPAQPRPKTPKSDDEDEEEDKDKEKKDGEED---EKKKGPSTLSKLTAEEAEHQALQAAV 350
K P PK + D EDEE +D++ GEE+ +KKG + + Q +AAV
Sbjct: 15 KKMAPPPKEVEEDSEDEEMSEDEDDSSGEEEVVIPQKKGKKATTTPAKKVVVSQTKKAAV 74
Score = 34.3 bits (77), Expect = 1.1
Identities = 13/23 (56%), Positives = 20/23 (86%)
Frame = -2
Query: 484 DDEDEEEDKDKEKKDGEEDEKKK 416
DDEDEEED+D+E ++ +EDE ++
Sbjct: 249 DDEDEEEDEDEEDEEDDEDEDEE 271
Score = 32.7 bits (73), Expect = 3.1
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 3/58 (5%)
Frame = -2
Query: 517 PAQPRPKTPKS---DDEDEEEDKDKEKKDGEEDEKKKGPSTLSKLTAEEAEHQALQAA 353
PA+ P KS ++ED+E+D+D+E+ + EEDE+ EE E + ++AA
Sbjct: 229 PAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEEDEEDDEDED------EEEEEEPVKAA 280
>pir||JH0148 nucleolin - rat
Length = 712
Score = 43.1 bits (100), Expect = 0.002
Identities = 20/59 (33%), Positives = 35/59 (58%)
Frame = -2
Query: 523 LKPAQPRPKTPKSDDEDEEEDKDKEKKDGEEDEKKKGPSTLSKLTAEEAEHQALQAAVL 347
+KPA+ P P S+DEDEE+D D++ D +E+E+++ S + A+ + A V+
Sbjct: 174 VKPAKAAPAAPASEDEDEEDDDDEDDDDDDEEEEEEDDSEEEVMEITPAKGKKTPAKVV 232
Score = 34.3 bits (77), Expect = 1.1
Identities = 20/60 (33%), Positives = 29/60 (48%), Gaps = 3/60 (5%)
Frame = -2
Query: 520 KPAQPRPKTPKSDDEDEEEDKDKEKKDGEED---EKKKGPSTLSKLTAEEAEHQALQAAV 350
K P PK + D EDEE +D++ GEE+ +KKG + + Q +AAV
Sbjct: 14 KKMAPPPKEVEEDSEDEEMSEDEDDSSGEEEVVIPQKKGKKATTTPAKKVVVSQTKKAAV 73
Score = 34.3 bits (77), Expect = 1.1
Identities = 13/23 (56%), Positives = 20/23 (86%)
Frame = -2
Query: 484 DDEDEEEDKDKEKKDGEEDEKKK 416
DDEDEEED+D+E ++ +EDE ++
Sbjct: 248 DDEDEEEDEDEEDEEDDEDEDEE 270
Score = 32.7 bits (73), Expect = 3.1
Identities = 20/58 (34%), Positives = 33/58 (56%), Gaps = 3/58 (5%)
Frame = -2
Query: 517 PAQPRPKTPKS---DDEDEEEDKDKEKKDGEEDEKKKGPSTLSKLTAEEAEHQALQAA 353
PA+ P KS ++ED+E+D+D+E+ + EEDE+ EE E + ++AA
Sbjct: 228 PAKVVPVKAKSVAEEEEDDEDDEDEEEDEDEEDEEDDEDED------EEEEEEPVKAA 279
>dbj|BAC34476.1| unnamed protein product [Mus musculus]
Length = 550
Score = 42.4 bits (98), Expect = 0.004
Identities = 18/47 (38%), Positives = 29/47 (61%)
Frame = -2
Query: 523 LKPAQPRPKTPKSDDEDEEEDKDKEKKDGEEDEKKKGPSTLSKLTAE 383
+KPA+ P P S+DE+++ED+D E+ D EE+E + TA+
Sbjct: 20 VKPAKAAPAAPASEDEEDDEDEDDEEDDDEEEEDDSEEEVMEITTAK 66
Score = 32.7 bits (73), Expect = 3.1
Identities = 20/58 (34%), Positives = 32/58 (54%), Gaps = 3/58 (5%)
Frame = -2
Query: 517 PAQPRPKTPKS---DDEDEEEDKDKEKKDGEEDEKKKGPSTLSKLTAEEAEHQALQAA 353
PA+ P KS +++DEEED+D E +D EE++ + EE E + ++AA
Sbjct: 71 PAKVVPMKAKSVAEEEDDEEEDEDDEDEDDEEEDDEDDDE-------EEEEEEPVKAA 121
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 476,530,157
Number of Sequences: 1393205
Number of extensions: 10872522
Number of successful extensions: 142358
Number of sequences better than 10.0: 1478
Number of HSP's better than 10.0 without gapping: 61630
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 108329
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20095422690
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)