Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
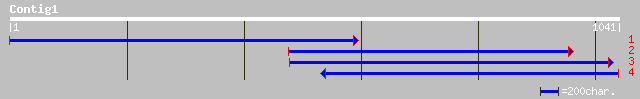
Query= KMC002745A_C01 KMC002745A_c01
(1032 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAF91323.1|AF244889_1 receptor-like protein kinase 2 [Glycine... 193 2e-48
gb|AAF91324.1|AF244890_1 receptor-like protein kinase 3 [Glycine... 189 5e-47
gb|AAF91322.1|AF244888_1 receptor-like protein kinase 1 [Glycine... 170 2e-41
gb|AAO26313.1| receptor-like protein kinase [Elaeis guineensis] 140 2e-32
ref|NP_201371.1| leucine-rich repeat transmembrane protein kinas... 126 2e-28
>gb|AAF91323.1|AF244889_1 receptor-like protein kinase 2 [Glycine max]
Length = 1012
Score = 193 bits (491), Expect(2) = 2e-48
Identities = 98/114 (85%), Positives = 107/114 (92%), Gaps = 3/114 (2%)
Frame = -2
Query: 1010 GVDIVQWVREMTDSNKEGVVKVLDPRLSSVPLHEVMHMFYVAILCVEEQAVERPTMREVV 831
GVDIVQWVR+MTDSNKEGV+KVLDPRL SVPLHEVMH+FYVA+LCVEEQAVERPTMREVV
Sbjct: 899 GVDIVQWVRKMTDSNKEGVLKVLDPRLPSVPLHEVMHVFYVAMLCVEEQAVERPTMREVV 958
Query: 830 QILTEL---PGSKQGDLTITESSLPSSNALESPTAASKDHENPPQSPPTDLLSI 678
QILTEL PGSK+GDLTITESSL SSNALESP++ASK+ +NPPQSPP DLLSI
Sbjct: 959 QILTELPKPPGSKEGDLTITESSLSSSNALESPSSASKEDQNPPQSPPPDLLSI 1012
Score = 22.3 bits (46), Expect(2) = 2e-48
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = -1
Query: 1032 PVGEFGDG 1009
PVGEFGDG
Sbjct: 892 PVGEFGDG 899
>gb|AAF91324.1|AF244890_1 receptor-like protein kinase 3 [Glycine max]
Length = 1012
Score = 189 bits (479), Expect(2) = 5e-47
Identities = 96/114 (84%), Positives = 106/114 (92%), Gaps = 3/114 (2%)
Frame = -2
Query: 1010 GVDIVQWVREMTDSNKEGVVKVLDPRLSSVPLHEVMHMFYVAILCVEEQAVERPTMREVV 831
GVDIVQWVR+MTDSNKEGV+KVLDPRL SVPLHEVMH+FYVA+LCVEEQAVERPTMREVV
Sbjct: 899 GVDIVQWVRKMTDSNKEGVLKVLDPRLPSVPLHEVMHVFYVAMLCVEEQAVERPTMREVV 958
Query: 830 QILTEL---PGSKQGDLTITESSLPSSNALESPTAASKDHENPPQSPPTDLLSI 678
QILTEL P SK+G+LTITESSL SSNALESP++ASK+ +NPPQSPP DLLSI
Sbjct: 959 QILTELPKPPDSKEGNLTITESSLSSSNALESPSSASKEDQNPPQSPPPDLLSI 1012
Score = 22.3 bits (46), Expect(2) = 5e-47
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = -1
Query: 1032 PVGEFGDG 1009
PVGEFGDG
Sbjct: 892 PVGEFGDG 899
>gb|AAF91322.1|AF244888_1 receptor-like protein kinase 1 [Glycine max]
Length = 1008
Score = 170 bits (431), Expect(2) = 2e-41
Identities = 89/114 (78%), Positives = 97/114 (85%), Gaps = 3/114 (2%)
Frame = -2
Query: 1010 GVDIVQWVREMTDSNKEGVVKVLDPRLSSVPLHEVMHMFYVAILCVEEQAVERPTMREVV 831
GVDIVQWVR+MTDSNKEGV+KVLD RL SVPLHEVMH+FYVA+LCVEEQAVERPTMREVV
Sbjct: 895 GVDIVQWVRKMTDSNKEGVLKVLDSRLPSVPLHEVMHVFYVAMLCVEEQAVERPTMREVV 954
Query: 830 QILTELPGSKQGDLTITESSLPSSNALESPTAAS---KDHENPPQSPPTDLLSI 678
QILTELP ITESSL SSN+L SPT AS KD+++PPQSPP DLLSI
Sbjct: 955 QILTELPKPPSSKHAITESSLSSSNSLGSPTTASKEPKDNQHPPQSPPPDLLSI 1008
Score = 22.3 bits (46), Expect(2) = 2e-41
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = -1
Query: 1032 PVGEFGDG 1009
PVGEFGDG
Sbjct: 888 PVGEFGDG 895
>gb|AAO26313.1| receptor-like protein kinase [Elaeis guineensis]
Length = 481
Score = 140 bits (352), Expect(2) = 2e-32
Identities = 74/113 (65%), Positives = 89/113 (78%), Gaps = 2/113 (1%)
Frame = -2
Query: 1010 GVDIVQWVREMTDSNKEGVVKVLDPRLSSVPLHEVMHMFYVAILCVEEQAVERPTMREVV 831
GVDIVQWVR++TD+NKEG++K++DPRLSSVPLHE MH+FYVA+LCVEEQ+VERPTMREVV
Sbjct: 374 GVDIVQWVRKVTDTNKEGILKIIDPRLSSVPLHEAMHVFYVAMLCVEEQSVERPTMREVV 433
Query: 830 QILTELPG--SKQGDLTITESSLPSSNALESPTAASKDHENPPQSPPTDLLSI 678
QILTELPG +KQG+ + P E+P SK + QSPP DLLSI
Sbjct: 434 QILTELPGTPAKQGE----DPPPPPPVGGETPPRESKPRQE-QQSPPPDLLSI 481
Score = 22.3 bits (46), Expect(2) = 2e-32
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = -1
Query: 1032 PVGEFGDG 1009
PVGEFGDG
Sbjct: 367 PVGEFGDG 374
>ref|NP_201371.1| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At5g65700.1, supported by cDNA: gi_20466695
[Arabidopsis thaliana] gi|7434429|pir||T05898
hypothetical protein F6H11.170 - Arabidopsis thaliana
gi|2827715|emb|CAA16688.1| receptor protein kinase - like
protein [Arabidopsis thaliana]
gi|10177328|dbj|BAB10677.1| receptor protein kinase-like
protein [Arabidopsis thaliana] gi|20466696|gb|AAM20665.1|
receptor protein kinase-like protein [Arabidopsis
thaliana]
Length = 1003
Score = 126 bits (317), Expect(2) = 2e-28
Identities = 66/95 (69%), Positives = 77/95 (80%), Gaps = 5/95 (5%)
Frame = -2
Query: 1010 GVDIVQWVREMTDSNKEGVVKVLDPRLSSVPLHEVMHMFYVAILCVEEQAVERPTMREVV 831
GVDIVQWVR+MTDSNK+ V+KVLDPRLSS+P+HEV H+FYVA+LCVEEQAVERPTMREVV
Sbjct: 903 GVDIVQWVRKMTDSNKDSVLKVLDPRLSSIPIHEVTHVFYVAMLCVEEQAVERPTMREVV 962
Query: 830 QILTE---LPGSKQGDLT--ITESSLPSSNALESP 741
QILTE LP SK +T ES L + ++SP
Sbjct: 963 QILTEIPKLPPSKDQPMTESAPESELSPKSGVQSP 997
Score = 22.3 bits (46), Expect(2) = 2e-28
Identities = 8/8 (100%), Positives = 8/8 (100%)
Frame = -1
Query: 1032 PVGEFGDG 1009
PVGEFGDG
Sbjct: 896 PVGEFGDG 903
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 899,615,036
Number of Sequences: 1393205
Number of extensions: 19765853
Number of successful extensions: 47002
Number of sequences better than 10.0: 453
Number of HSP's better than 10.0 without gapping: 44674
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46936
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 60429070113
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)