Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002734A_C01 KMC002734A_c01
(836 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
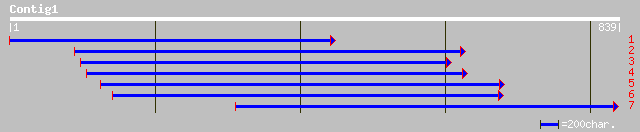
ref|NP_174656.1| hypothetical protein; protein id: At1g33940.1 [... 161 1e-38
ref|NP_197371.1| putative protein; protein id: At5g18700.1 [Arab... 154 2e-36
dbj|BAB03430.1| unnamed protein product [Oryza sativa (japonica ... 136 3e-31
ref|NP_498160.1| RNB-like protein [Caenorhabditis elegans] gi|13... 33 5.2
gb|AAL56654.1| spike glycoprotein S1 subunit [avian infectious b... 33 6.8
>ref|NP_174656.1| hypothetical protein; protein id: At1g33940.1 [Arabidopsis
thaliana] gi|25373112|pir||C86463 hypothetical protein
T3M13.4 - Arabidopsis thaliana
gi|12324512|gb|AAG52217.1|AC022288_16 hypothetical
protein; 18034-19895 [Arabidopsis thaliana]
Length = 497
Score = 161 bits (407), Expect = 1e-38
Identities = 88/147 (59%), Positives = 108/147 (72%), Gaps = 7/147 (4%)
Frame = -3
Query: 816 DPQQGIKDIADFGSNLGVLLE---LSSYAETSIADIASECVVLLLKAAPREGTTGLLTNL 646
DPQ+ I+DIADFGSN+G+ L L ++ADIASECVVLLLKAA RE TTG LTNL
Sbjct: 351 DPQECIRDIADFGSNIGLFLHFAGLDDDTSIAVADIASECVVLLLKAASREATTGFLTNL 410
Query: 645 PKVSVILESWSKGTP---HLMV-QRMLHALGYACKQYLLHAMILSISIPEISRIEVIVSD 478
PK++ IL+SW + HL+V +R+LH LGYACKQYL AMILSIS +IS+I IVS+
Sbjct: 411 PKITQILDSWRRRKSTELHLLVLKRVLHCLGYACKQYLSQAMILSISGHDISKINAIVSE 470
Query: 477 LKSSGVPALAKTAGLAALELQRLPRCI 397
+K+S V L A L A+ELQRLPRC+
Sbjct: 471 MKNSDVAGLNSVASLVAMELQRLPRCV 497
>ref|NP_197371.1| putative protein; protein id: At5g18700.1 [Arabidopsis thaliana]
Length = 1317
Score = 154 bits (389), Expect = 2e-36
Identities = 85/145 (58%), Positives = 105/145 (71%), Gaps = 7/145 (4%)
Frame = -3
Query: 816 DPQQGIKDIADFGSNLGVLLE---LSSYAETSIADIASECVVLLLKAAPREGTTGLLTNL 646
DPQ+ I+DIADFGSN+G+ L L ++ADIASECVVLLLKAA RE TTG LTNL
Sbjct: 1173 DPQECIRDIADFGSNIGLFLHFAGLDDDTSIAVADIASECVVLLLKAASREATTGFLTNL 1232
Query: 645 PKVSVILESWSKGTP---HLMV-QRMLHALGYACKQYLLHAMILSISIPEISRIEVIVSD 478
PK++ IL+SW + HL+V +R+LH LGYACKQYL AMILSIS ++S+I IVS+
Sbjct: 1233 PKITPILDSWRRRKSTELHLLVLKRVLHCLGYACKQYLSQAMILSISGHDVSKINAIVSE 1292
Query: 477 LKSSGVPALAKTAGLAALELQRLPR 403
+K+S L A L A+ELQRLPR
Sbjct: 1293 MKNSDAAGLNSIASLVAMELQRLPR 1317
>dbj|BAB03430.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
gi|9711909|dbj|BAB08000.1| P0462H08.23 [Oryza sativa
(japonica cultivar-group)]
Length = 185
Score = 136 bits (343), Expect = 3e-31
Identities = 70/139 (50%), Positives = 94/139 (67%)
Frame = -3
Query: 816 DPQQGIKDIADFGSNLGVLLELSSYAETSIADIASECVVLLLKAAPREGTTGLLTNLPKV 637
D Q + DI DFG N+G+ L+L ++ I+D+AS+C+VLLLKAAPRE T GLLTNLPK+
Sbjct: 47 DQQSCVMDICDFGGNMGIFLDLVGSSDPHISDLASDCLVLLLKAAPREATVGLLTNLPKL 106
Query: 636 SVILESWSKGTPHLMVQRMLHALGYACKQYLLHAMILSISIPEISRIEVIVSDLKSSGVP 457
SV+L+ GT L + R+L+ L ++C+QYL MI+SIS+ + R+E +VS K S
Sbjct: 107 SVVLDLLKHGT-CLRLTRLLYCLAFSCRQYLAQGMIVSISLSALMRVEALVSAFKGSHDG 165
Query: 456 ALAKTAGLAALELQRLPRC 400
LA A ELQRLPRC
Sbjct: 166 CLADAASYLGAELQRLPRC 184
>ref|NP_498160.1| RNB-like protein [Caenorhabditis elegans]
gi|1353148|sp|Q09568|YR86_CAEEL Hypothetical protein
F48E8.6 in chromosome III gi|7503759|pir||T16409
hypothetical protein F48E8.6 - Caenorhabditis elegans
gi|746490|gb|AAC46543.1| Hypothetical protein F48E8.6
[Caenorhabditis elegans]
Length = 817
Score = 33.1 bits (74), Expect = 5.2
Identities = 23/97 (23%), Positives = 44/97 (44%), Gaps = 7/97 (7%)
Frame = -3
Query: 540 HAMILSISIPEISRIEVIVSDLKSSGVPALAKTAGLAALELQ------RLPRCI*IIVVS 379
HA++ + P+ I+ + G P +T+GL + L+ RL CI ++ S
Sbjct: 540 HALLRNHPPPKEKMIKDVAEQCARIGFPLDGRTSGLLSTSLRKYQGKSRLDMCIRQVISS 599
Query: 378 ILLSSIRLPRCICIFYLYCSCL-KFKIKVEHFGEFHS 271
+ + ++ + C F + S F + V+H+ F S
Sbjct: 600 LTIKPMQQAKYFCTFEMPLSFYHHFALNVDHYTHFTS 636
>gb|AAL56654.1| spike glycoprotein S1 subunit [avian infectious bronchitis virus]
Length = 302
Score = 32.7 bits (73), Expect = 6.8
Identities = 22/59 (37%), Positives = 28/59 (47%), Gaps = 4/59 (6%)
Frame = +3
Query: 315 DMNNKGRRCRCSEAAEYWTTEY*QQLFKCSEAAAAIPRLPTQ----QFWLKQEHLNFSD 479
+ NN G R RC+ A +W+ F S AA P L + QFW + H NFSD
Sbjct: 55 EYNNAGGRPRCTLGAIHWSKN-----FSASSAAMTAPLLGMEWSPNQFW--RLHCNFSD 106
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 678,476,594
Number of Sequences: 1393205
Number of extensions: 14044201
Number of successful extensions: 31192
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 30199
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31170
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 43480044972
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)