Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002727A_C01 KMC002727A_c01
(548 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
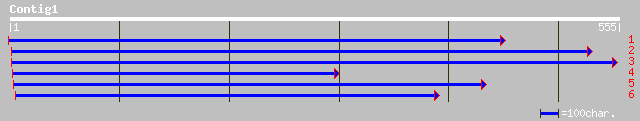
dbj|BAB63568.1| putative helicase-like transcription factor [Ory... 103 1e-21
pir||D96540 hypothetical protein F11F12.23 [imported] - Arabidop... 103 2e-21
ref|NP_564568.1| DNA-binding protein, putative; protein id: At1g... 103 2e-21
ref|NP_176309.1| hypothetical protein; protein id: At1g61140.1 [... 101 5e-21
ref|NP_188635.1| uteroglobin promoter-binding protein, RUSH-1alp... 99 3e-20
>dbj|BAB63568.1| putative helicase-like transcription factor [Oryza sativa (japonica
cultivar-group)]
Length = 1082
Score = 103 bits (258), Expect = 1e-21
Identities = 51/64 (79%), Positives = 59/64 (91%)
Frame = -3
Query: 546 RAHRIGQTRPVTVLRLTVRDTVEDRILALQPKKRKMVSSAFGEDGSGSGQSRLTVDDLKY 367
RAHRIGQTRPVTV RLT++DTVEDRILALQ KKR+MV+SAFGED SG+ Q+RLTV+DL Y
Sbjct: 1019 RAHRIGQTRPVTVSRLTIKDTVEDRILALQEKKREMVASAFGEDKSGAHQTRLTVEDLNY 1078
Query: 366 LFMM 355
LFM+
Sbjct: 1079 LFMV 1082
>pir||D96540 hypothetical protein F11F12.23 [imported] - Arabidopsis thaliana
gi|9454567|gb|AAF87890.1|AC012561_23 Similar tp
transcription factors [Arabidopsis thaliana]
Length = 1062
Score = 103 bits (256), Expect = 2e-21
Identities = 50/64 (78%), Positives = 58/64 (90%)
Frame = -3
Query: 546 RAHRIGQTRPVTVLRLTVRDTVEDRILALQPKKRKMVSSAFGEDGSGSGQSRLTVDDLKY 367
RAHRIGQTRPVTV R+T+++TVEDRILALQ +KRKMV+SAFGED GS +RLTVDDLKY
Sbjct: 999 RAHRIGQTRPVTVTRITIKNTVEDRILALQEEKRKMVASAFGEDHGGSSATRLTVDDLKY 1058
Query: 366 LFMM 355
LFM+
Sbjct: 1059 LFMV 1062
>ref|NP_564568.1| DNA-binding protein, putative; protein id: At1g50410.1, supported by
cDNA: gi_14532629 [Arabidopsis thaliana]
gi|14532630|gb|AAK64043.1| putative DNA-binding protein
[Arabidopsis thaliana] gi|23296945|gb|AAN13207.1|
putative DNA-binding protein [Arabidopsis thaliana]
Length = 981
Score = 103 bits (256), Expect = 2e-21
Identities = 50/64 (78%), Positives = 58/64 (90%)
Frame = -3
Query: 546 RAHRIGQTRPVTVLRLTVRDTVEDRILALQPKKRKMVSSAFGEDGSGSGQSRLTVDDLKY 367
RAHRIGQTRPVTV R+T+++TVEDRILALQ +KRKMV+SAFGED GS +RLTVDDLKY
Sbjct: 918 RAHRIGQTRPVTVTRITIKNTVEDRILALQEEKRKMVASAFGEDHGGSSATRLTVDDLKY 977
Query: 366 LFMM 355
LFM+
Sbjct: 978 LFMV 981
>ref|NP_176309.1| hypothetical protein; protein id: At1g61140.1 [Arabidopsis thaliana]
Length = 1287
Score = 101 bits (252), Expect = 5e-21
Identities = 51/63 (80%), Positives = 55/63 (86%)
Frame = -3
Query: 546 RAHRIGQTRPVTVLRLTVRDTVEDRILALQPKKRKMVSSAFGEDGSGSGQSRLTVDDLKY 367
RAHRIGQTRPVTV+R TV+DTVEDRILALQ KKR MV+SAFGED GS QS LTV+DL Y
Sbjct: 1222 RAHRIGQTRPVTVVRFTVKDTVEDRILALQQKKRMMVASAFGEDEKGSRQSHLTVEDLSY 1281
Query: 366 LFM 358
LFM
Sbjct: 1282 LFM 1284
>ref|NP_188635.1| uteroglobin promoter-binding protein, RUSH-1alpha-like, putative;
protein id: At3g20010.1 [Arabidopsis thaliana]
gi|11994776|dbj|BAB03166.1| transcription factor-like
protein [Arabidopsis thaliana]
Length = 1047
Score = 99.4 bits (246), Expect = 3e-20
Identities = 48/64 (75%), Positives = 56/64 (87%)
Frame = -3
Query: 546 RAHRIGQTRPVTVLRLTVRDTVEDRILALQPKKRKMVSSAFGEDGSGSGQSRLTVDDLKY 367
RAHRIGQTRPVTV R+T++DTVEDRIL LQ +KR MV+SAFGE+ GS +RLTVDDLKY
Sbjct: 984 RAHRIGQTRPVTVTRITIKDTVEDRILKLQEEKRTMVASAFGEEHGGSSATRLTVDDLKY 1043
Query: 366 LFMM 355
LFM+
Sbjct: 1044 LFMV 1047
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 428,579,105
Number of Sequences: 1393205
Number of extensions: 8328604
Number of successful extensions: 19689
Number of sequences better than 10.0: 433
Number of HSP's better than 10.0 without gapping: 18982
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19659
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 18947112822
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)