Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002720A_C01 KMC002720A_c01
(594 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
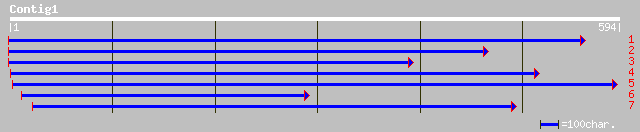
dbj|BAB09286.1| receptor protein kinase-like protein [Arabidopsi... 97 2e-19
pir||T05050 protein kinase homolog M3E9.30 - Arabidopsis thalian... 94 1e-18
gb|AAL32011.1|AF436829_1 AT4g26540/M3E9_30 [Arabidopsis thaliana] 84 1e-15
emb|CAC20842.1| receptor protein kinase [Pinus sylvestris] 50 3e-05
ref|NP_189066.1| leucine-rich repeat transmembrane protein kinas... 49 5e-05
>dbj|BAB09286.1| receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 1015
Score = 97.1 bits (240), Expect = 2e-19
Identities = 51/85 (60%), Positives = 62/85 (72%), Gaps = 4/85 (4%)
Frame = -2
Query: 593 MHEMLQTLAVSFLCVSTRAGDRPTMKDIVAMLKEIKPVETSRGENDKLKGGFTA--HSSP 420
MHEMLQTLAVSFLCVS +A DRP MKDIVAMLKEI+ + R E+D +KGG P
Sbjct: 931 MHEMLQTLAVSFLCVSNKASDRPMMKDIVAMLKEIRQFDMDRSESDMIKGGKCEKWQPQP 990
Query: 419 PPPKNVVS--HGSSSCSYNFSDDSV 351
PP+ +VS GSS+CS+ +SD+SV
Sbjct: 991 LPPEKIVSTPRGSSNCSFAYSDESV 1015
>pir||T05050 protein kinase homolog M3E9.30 - Arabidopsis thaliana
gi|2982452|emb|CAA18216.1| receptor protein kinase-like
protein [Arabidopsis thaliana] gi|7269506|emb|CAB79509.1|
receptor protein kinase-like protein [Arabidopsis
thaliana]
Length = 1029
Score = 94.0 bits (232), Expect = 1e-18
Identities = 47/85 (55%), Positives = 59/85 (69%), Gaps = 4/85 (4%)
Frame = -2
Query: 593 MHEMLQTLAVSFLCVSTRAGDRPTMKDIVAMLKEIKPVETSRGENDKLKGGFTAHSSPPP 414
MHEMLQTLAV+FLCVS +A +RP MKD+VAML EI+ ++ R E +K+K G P
Sbjct: 945 MHEMLQTLAVAFLCVSNKANERPLMKDVVAMLTEIRHIDVGRSETEKIKAGGCGSKEPQQ 1004
Query: 413 ----PKNVVSHGSSSCSYNFSDDSV 351
K + SHGSS+CS+ FSDDSV
Sbjct: 1005 FMSNEKIINSHGSSNCSFAFSDDSV 1029
>gb|AAL32011.1|AF436829_1 AT4g26540/M3E9_30 [Arabidopsis thaliana]
Length = 1096
Score = 84.0 bits (206), Expect = 1e-15
Identities = 42/80 (52%), Positives = 53/80 (65%), Gaps = 4/80 (5%)
Frame = -2
Query: 593 MHEMLQTLAVSFLCVSTRAGDRPTMKDIVAMLKEIKPVETSRGENDKLKGGFTAHSSPPP 414
MHEMLQTLAV+FLCVS +A +RP MKD+VAML EI+ ++ R E +K+K G P
Sbjct: 1007 MHEMLQTLAVAFLCVSNKANERPLMKDVVAMLTEIRHIDVGRSETEKIKAGGCGSKEPQQ 1066
Query: 413 ----PKNVVSHGSSSCSYNF 366
K + SHGSS+CS F
Sbjct: 1067 FMSNEKIINSHGSSNCSLRF 1086
>emb|CAC20842.1| receptor protein kinase [Pinus sylvestris]
Length = 1145
Score = 49.7 bits (117), Expect = 3e-05
Identities = 33/86 (38%), Positives = 45/86 (51%), Gaps = 4/86 (4%)
Frame = -2
Query: 593 MHEMLQTLAVSFLCVSTRAGDRPTMKDIVAMLKEIK-PVETSRGENDKLKGGFTAHS--- 426
+ EMLQ L V+FLCV++ +RPTMKD+ A+LKEI+ G+ D L A
Sbjct: 1045 IQEMLQVLGVAFLCVNSNPDERPTMKDVAALLKEIRHDCHDYNGKADLLLKQTPAPGSTR 1104
Query: 425 SPPPPKNVVSHGSSSCSYNFSDDSVS 348
SP P + S SS +S S +
Sbjct: 1105 SPNPTADARSPVGSSFGLEYSSASTT 1130
>ref|NP_189066.1| leucine-rich repeat transmembrane protein kinase, putative; protein
id: At3g24240.1 [Arabidopsis thaliana]
Length = 1141
Score = 48.9 bits (115), Expect = 5e-05
Identities = 26/58 (44%), Positives = 33/58 (56%)
Frame = -2
Query: 587 EMLQTLAVSFLCVSTRAGDRPTMKDIVAMLKEIKPVETSRGENDKLKGGFTAHSSPPP 414
EM+Q L + LCV++ +RPTMKD+ AMLKEIK + D L SPPP
Sbjct: 1034 EMMQVLGTALLCVNSSPDERPTMKDVAAMLKEIKQEREEYAKVDLL-----LKKSPPP 1086
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 450,679,418
Number of Sequences: 1393205
Number of extensions: 8965507
Number of successful extensions: 21245
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 20289
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 21204
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22854740960
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)