Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002710A_C01 KMC002710A_c01
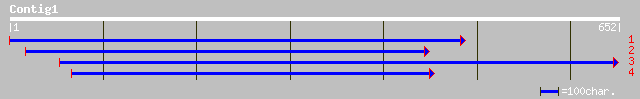
(649 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_191562.1| squamosa promoter binding protein-like 12; prot... 99 5e-20
pir||T52602 squamosa promoter binding protein 1 [imported] - Ara... 97 1e-19
ref|NP_182230.2| putative squamosa-promoter binding protein; pro... 97 1e-19
ref|NP_182231.1| unknown protein [Arabidopsis thaliana] gi|74524... 97 1e-19
dbj|BAA96636.1| unnamed protein product [Oryza sativa (japonica ... 60 3e-08
>ref|NP_191562.1| squamosa promoter binding protein-like 12; protein id: At3g60030.1
[Arabidopsis thaliana] gi|11358858|pir||T47827 squamosa
promoter binding protein-like 12 [imported] - Arabidopsis
thaliana gi|6006395|emb|CAB56768.1| squamosa promoter
binding protein-like 12 [Arabidopsis thaliana]
gi|6006403|emb|CAB56769.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|7076756|emb|CAB75918.1| squamosa promoter binding
protein-like 12 [Arabidopsis thaliana]
gi|28392866|gb|AAO41870.1| putative squamosa promoter
binding protein 12 [Arabidopsis thaliana]
Length = 927
Score = 99.0 bits (245), Expect = 5e-20
Identities = 50/92 (54%), Positives = 64/92 (69%)
Frame = -2
Query: 639 FNANEXQDELLSTSFEIGKAEVKSVQKLCKLCDLKLSCRTAAGRSLVYKPAMLSMVAVAA 460
FN Q++ + + E+ + + CKLCD K T +S+ Y+PAMLSMVA+AA
Sbjct: 839 FNIEHKQEK--RSPMDSSSLEITQINQ-CKLCDHKRVFVTTHHKSVAYRPAMLSMVAIAA 895
Query: 459 VCVCVALLFKSSPEVLYIFRPFRWESLEFGTS 364
VCVCVALLFKS PEVLY+F+PFRWE LE+GTS
Sbjct: 896 VCVCVALLFKSCPEVLYVFQPFRWELLEYGTS 927
>pir||T52602 squamosa promoter binding protein 1 [imported] - Arabidopsis thaliana
gi|5931653|emb|CAB56580.1| squamosa promoter binding
protein-like 1 [Arabidopsis thaliana]
Length = 881
Score = 97.4 bits (241), Expect = 1e-19
Identities = 47/64 (73%), Positives = 54/64 (83%)
Frame = -2
Query: 555 CKLCDLKLSCRTAAGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYIFRPFRWESLE 376
CKLCD KL T RS+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PFRWE L+
Sbjct: 819 CKLCDHKLVYGTTR-RSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLD 877
Query: 375 FGTS 364
+GTS
Sbjct: 878 YGTS 881
>ref|NP_182230.2| putative squamosa-promoter binding protein; protein id: At2g47070.1
[Arabidopsis thaliana] gi|25456637|pir||T52601 squamosa
promoter binding protein 1 [imported] - Arabidopsis
thaliana gi|3650274|emb|CAA09698.1| squamosa-promoter
binding protein-like 1 [Arabidopsis thaliana]
gi|5931655|emb|CAB56581.1| squamosa promoter binding
protein-like 1 [Arabidopsis thaliana]
Length = 881
Score = 97.4 bits (241), Expect = 1e-19
Identities = 47/64 (73%), Positives = 54/64 (83%)
Frame = -2
Query: 555 CKLCDLKLSCRTAAGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYIFRPFRWESLE 376
CKLCD KL T RS+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PFRWE L+
Sbjct: 819 CKLCDHKLVYGTTR-RSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLD 877
Query: 375 FGTS 364
+GTS
Sbjct: 878 YGTS 881
>ref|NP_182231.1| unknown protein [Arabidopsis thaliana] gi|7452429|pir||T02179
hypothetical protein At2g47080 [imported] - Arabidopsis
thaliana gi|3522938|gb|AAC34220.1| unknown protein
[Arabidopsis thaliana]
Length = 383
Score = 97.4 bits (241), Expect = 1e-19
Identities = 47/64 (73%), Positives = 54/64 (83%)
Frame = -2
Query: 555 CKLCDLKLSCRTAAGRSLVYKPAMLSMVAVAAVCVCVALLFKSSPEVLYIFRPFRWESLE 376
CKLCD KL T RS+ Y+PAMLSMVA+AAVCVCVALLFKS PEVLY+F+PFRWE L+
Sbjct: 321 CKLCDHKLVYGTTR-RSVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLD 379
Query: 375 FGTS 364
+GTS
Sbjct: 380 YGTS 383
>dbj|BAA96636.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 862
Score = 59.7 bits (143), Expect = 3e-08
Identities = 32/80 (40%), Positives = 48/80 (60%), Gaps = 2/80 (2%)
Frame = -2
Query: 603 TSFEIGKAEVKSVQK--LCKLCDLKLSCRTAAGRSLVYKPAMLSMVAVAAVCVCVALLFK 430
T+F++ K + S + C+ C +L+ R R L +PA+LS+VA+AAVCVCV L+ +
Sbjct: 781 TAFDVEKGQQISTKPPLSCRQCLPELAYRHHLNRFLSTRPAVLSLVAIAAVCVCVGLIMQ 840
Query: 429 SSPEVLYIFRPFRWESLEFG 370
P + + PFRW SL G
Sbjct: 841 GPPHIGGMRGPFRWNSLRSG 860
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 538,711,456
Number of Sequences: 1393205
Number of extensions: 11677139
Number of successful extensions: 32404
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 30408
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 32262
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27576232529
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)