Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002709A_C01 KMC002709A_c01
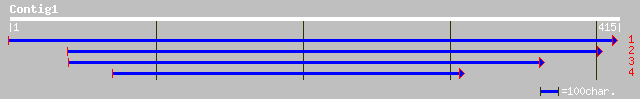
(410 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T02003 probable DNA-binding protein T15B16.12 - Arabidopsis... 82 2e-15
ref|NP_192081.1| WRKY family transcription factor; protein id: A... 82 2e-15
pir||T49114 hypothetical protein AT4g22070 - Arabidopsis thalian... 52 1e-06
ref|NP_567644.1| WRKY family transcription factor; protein id: A... 52 1e-06
dbj|BAA96574.1| unnamed protein product [Oryza sativa (japonica ... 52 2e-06
>pir||T02003 probable DNA-binding protein T15B16.12 - Arabidopsis thaliana
gi|3859603|gb|AAC72869.1| contains similarity to wild
oat DNA-binding protein ABF2 (GB:Z48431) [Arabidopsis
thaliana]
Length = 403
Score = 82.0 bits (201), Expect = 2e-15
Identities = 44/76 (57%), Positives = 57/76 (74%), Gaps = 5/76 (6%)
Frame = -2
Query: 403 QRPPT--VETMSAAIASDPNFTAALAAAISSIIRGSDGHNNSSSDVNNN---SNNGSAII 239
Q PP V+++ AAIA DPNFTAALAAAIS+II G + NN+++D+N+N + +G +
Sbjct: 328 QAPPREMVDSVRAAIAMDPNFTAALAAAISNIIGGGNNDNNNNTDINDNKVDAKSGGSSN 387
Query: 238 PGSPQLPQSCTTFSTN 191
SPQLPQSCTTFSTN
Sbjct: 388 GDSPQLPQSCTTFSTN 403
>ref|NP_192081.1| WRKY family transcription factor; protein id: At4g01720.1,
supported by cDNA: gi_19172391 [Arabidopsis thaliana]
gi|20978798|sp|Q9ZSI7|WR47_ARATH Probable WRKY
transcription factor 47 (WRKY DNA-binding protein 47)
gi|25407124|pir||B85022 probable DNA-binding protein
[imported] - Arabidopsis thaliana
gi|7268215|emb|CAB77742.1| putative DNA-binding protein
[Arabidopsis thaliana]
gi|19172392|gb|AAL85881.1|AF480165_1 WRKY transcription
factor 47 [Arabidopsis thaliana]
Length = 489
Score = 82.0 bits (201), Expect = 2e-15
Identities = 44/76 (57%), Positives = 57/76 (74%), Gaps = 5/76 (6%)
Frame = -2
Query: 403 QRPPT--VETMSAAIASDPNFTAALAAAISSIIRGSDGHNNSSSDVNNN---SNNGSAII 239
Q PP V+++ AAIA DPNFTAALAAAIS+II G + NN+++D+N+N + +G +
Sbjct: 414 QAPPREMVDSVRAAIAMDPNFTAALAAAISNIIGGGNNDNNNNTDINDNKVDAKSGGSSN 473
Query: 238 PGSPQLPQSCTTFSTN 191
SPQLPQSCTTFSTN
Sbjct: 474 GDSPQLPQSCTTFSTN 489
>pir||T49114 hypothetical protein AT4g22070 - Arabidopsis thaliana
gi|2961352|emb|CAA18110.1| putative protein [Arabidopsis
thaliana] gi|7269052|emb|CAB79162.1| putative protein
[Arabidopsis thaliana]
Length = 458
Score = 52.4 bits (124), Expect = 1e-06
Identities = 31/51 (60%), Positives = 37/51 (71%), Gaps = 3/51 (5%)
Frame = -2
Query: 391 TVETMSAAIASDPNFTAALAAAISSIIRGSDGHNNSSSDVNNN---SNNGS 248
+V SAAIASDPNF AALAAAI+SI+ GS NN+++ NNN SNN S
Sbjct: 408 SVSAASAAIASDPNFAAALAAAITSIMNGSSHQNNNTN--NNNVATSNNDS 456
>ref|NP_567644.1| WRKY family transcription factor; protein id: At4g22070.1,
supported by cDNA: gi_15990589 [Arabidopsis thaliana]
gi|20978775|sp|Q93WT0|WR31_ARATH Probable WRKY
transcription factor 31 (WRKY DNA-binding protein 31)
gi|15990590|gb|AAL11009.1| WRKY transcription factor 31
[Arabidopsis thaliana]
Length = 538
Score = 52.4 bits (124), Expect = 1e-06
Identities = 31/51 (60%), Positives = 37/51 (71%), Gaps = 3/51 (5%)
Frame = -2
Query: 391 TVETMSAAIASDPNFTAALAAAISSIIRGSDGHNNSSSDVNNN---SNNGS 248
+V SAAIASDPNF AALAAAI+SI+ GS NN+++ NNN SNN S
Sbjct: 488 SVSAASAAIASDPNFAAALAAAITSIMNGSSHQNNNTN--NNNVATSNNDS 536
>dbj|BAA96574.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 594
Score = 52.0 bits (123), Expect = 2e-06
Identities = 27/68 (39%), Positives = 42/68 (61%), Gaps = 10/68 (14%)
Frame = -2
Query: 403 QRPPTVETMSAAIASDPNFTAALAAAISSIIRGSDGHNNSSS----------DVNNNSNN 254
++P +ET++AA+A+DPNFT ALAAAISS++ G H S+ D N N ++
Sbjct: 499 RQPSVMETVTAALAADPNFTTALAAAISSVVAGGAHHQALSTTPRGSAAGAGDGNGNGSS 558
Query: 253 GSAIIPGS 230
+A+ G+
Sbjct: 559 AAAVATGA 566
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 340,890,299
Number of Sequences: 1393205
Number of extensions: 6888401
Number of successful extensions: 70364
Number of sequences better than 10.0: 195
Number of HSP's better than 10.0 without gapping: 22371
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 54388
length of database: 448,689,247
effective HSP length: 112
effective length of database: 292,650,287
effective search space used: 7023606888
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)