Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
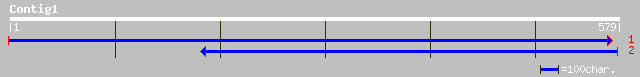
Query= KMC002699A_C01 KMC002699A_c01
(574 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_176175.1| nucleoporin 98-like protein; protein id: At1g59... 164 1e-39
ref|NP_172510.2| unknown protein; protein id: At1g10390.1, suppo... 162 2e-39
pir||C96620 protein T30E16.23 [imported] - Arabidopsis thaliana ... 155 3e-37
pir||H86237 protein F14N23.29 [imported] - Arabidopsis thaliana ... 139 3e-32
emb|CAB53357.1| nucleoporin CAN [Xenopus laevis] 74 1e-12
>ref|NP_176175.1| nucleoporin 98-like protein; protein id: At1g59660.1 [Arabidopsis
thaliana]
Length = 997
Score = 164 bits (414), Expect = 1e-39
Identities = 98/165 (59%), Positives = 117/165 (70%), Gaps = 6/165 (3%)
Frame = +3
Query: 93 MFGSTN--PLGQQS-SSPFGS-SQSVFGQ--NNSSSNPLAPKPFGSNTTPFGAQTGTSMC 254
MFGS+N P GQ S SSPFG+ + S+FGQ NN+S+NP A KPFG++T PFGAQTG+SM
Sbjct: 1 MFGSSNNNPFGQSSISSPFGTQTHSLFGQTNNNASNNPFATKPFGTST-PFGAQTGSSMF 59
Query: 255 GGTSTGVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVF 434
GGTSTGV GA Q SSPF +S AFGSS A G++STP+FG SS+S FGG+S F
Sbjct: 60 GGTSTGVFGAPQTSSPF-------GASPQAFGSSTQAFGASSTPSFG-SSNSPFGGTSTF 111
Query: 435 GQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
GQK FG + Q+SPFG Q SQP FG+S FGS T FGAS+
Sbjct: 112 GQK----SFGLSTPQSSPFGSTTQQSQPAFGNSTFGSSTPFGAST 152
Score = 90.5 bits (223), Expect = 1e-17
Identities = 70/167 (41%), Positives = 97/167 (57%), Gaps = 9/167 (5%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQS-VFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTG 272
FGS+ P G ++ FG+S + FG +N+S FG+ TP T T+ GG+ST
Sbjct: 142 FGSSTPFGASTTPAFGASSTPAFGVSNTSG-------FGATNTPGFGATNTTGFGGSSTP 194
Query: 273 VLGAAQQSSPFSSTT-AFGASSSPAFG-SSAPALGSTSTPTFGNSSSSSFGGSSVFGQKP 446
GA+ + S+ T AFGASS+P FG SS+PA G++ P FG+S ++ G++ F
Sbjct: 195 GFGASSTPAFGSTNTPAFGASSTPLFGSSSSPAFGASPAPAFGSSGNAF--GNNTFSSGG 252
Query: 447 AWGGFGSTPT----QTSPFGGAAQPSQPGFGSS-IFG-SPTAFGASS 569
A+G STPT TS FG ++ PS FGSS FG S +AFG+SS
Sbjct: 253 AFGS-SSTPTFGASNTSAFGASSSPSF-NFGSSPAFGQSTSAFGSSS 297
Score = 63.9 bits (154), Expect = 1e-09
Identities = 53/136 (38%), Positives = 68/136 (49%), Gaps = 22/136 (16%)
Frame = +3
Query: 96 FGSTN---------PLGQQSSSP---------FGSSQSVFGQNNSSSNPLAPKPFGSNTT 221
FGSTN PL SSSP FGSS + FG N SS FGS++T
Sbjct: 204 FGSTNTPAFGASSTPLFGSSSSPAFGASPAPAFGSSGNAFGNNTFSSGGA----FGSSST 259
Query: 222 PFGAQTGTSMCGGTSTGVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFG-- 395
P + TS G +S+ + ST+AFG+SS FGS+ +LGST +P FG
Sbjct: 260 PTFGASNTSAFGASSSPSFNFGSSPAFGQSTSAFGSSS---FGSTQSSLGSTPSP-FGAQ 315
Query: 396 --NSSSSSFGGSSVFG 437
+S+S+FGG S G
Sbjct: 316 GAQASTSTFGGQSTIG 331
Score = 62.8 bits (151), Expect = 3e-09
Identities = 57/161 (35%), Positives = 79/161 (48%), Gaps = 6/161 (3%)
Frame = +3
Query: 102 STNPLGQQSSSPFGSSQS-VFGQNNSSSNPLAPKP-FGSNTTPFGAQTGTS--MCGGTST 269
ST G ++ FG+S + +FG ++S + +P P FGS+ FG T +S G +ST
Sbjct: 200 STPAFGSTNTPAFGASSTPLFGSSSSPAFGASPAPAFGSSGNAFGNNTFSSGGAFGSSST 259
Query: 270 GVLGAAQQSS-PFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQKP 446
GA+ S+ SS+ +F SSPAFG S A GS+S FG++ SS G P
Sbjct: 260 PTFGASNTSAFGASSSPSFNFGSSPAFGQSTSAFGSSS---FGSTQSS-------LGSTP 309
Query: 447 AWGGFGSTPTQTSPFGGAAQ-PSQPGFGSSIFGSPTAFGAS 566
+ G TS FGG + Q G I +PT AS
Sbjct: 310 SPFGAQGAQASTSTFGGQSTIGGQQGGSRVIPYAPTTDTAS 350
Score = 60.1 bits (144), Expect = 2e-08
Identities = 58/181 (32%), Positives = 81/181 (44%), Gaps = 34/181 (18%)
Frame = +3
Query: 102 STNPLGQQSSSPFGSS----QSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTST 269
+T GQ ++ F S+ SVFG ++S + + +P GS+ FG+ G S
Sbjct: 447 TTPSFGQPTTPSFRSTVSNTTSVFGSSSSLTTNTS-QPLGSSI--FGSTPAHGSTPGFSI 503
Query: 270 GVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQ--K 443
G +Q S F S +F +++PAF ++P G +TP G S SSVFGQ
Sbjct: 504 GGFNNSQSSPLFGSNPSFAQNTTPAFSQTSPLFGQNTTPALGQS-------SSVFGQNTN 556
Query: 444 PAW----------GGFGST-------PTQTSPFG---------GAAQPSQP--GFGSSIF 539
PA GFG+T T SPFG +AQP+QP FG + F
Sbjct: 557 PALVQSNTFSTPSTGFGNTFSSSSSLTTSISPFGQITPAVTPFQSAQPTQPLGAFGFNNF 616
Query: 540 G 542
G
Sbjct: 617 G 617
Score = 50.8 bits (120), Expect = 1e-05
Identities = 61/205 (29%), Positives = 78/205 (37%), Gaps = 47/205 (22%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTGV 275
FGS+ GQ S+S FGSS FG SS P PFG+ GAQ TS GG ST
Sbjct: 280 FGSSPAFGQ-STSAFGSSS--FGSTQSSLGS-TPSPFGAQ----GAQASTSTFGGQST-- 329
Query: 276 LGAAQQSS---PFSSTT--AFGASSSPAFGSSAPALGSTSTPTFGNSSSSSF-----GGS 425
+G Q S P++ TT A G S S A+ + + GG
Sbjct: 330 IGGQQGGSRVIPYAPTTDTASGTESKSERLQSISAMPAHKGKNMEELRWEDYQRGDKGGQ 389
Query: 426 SVFGQKPAWGGFGSTPTQ-----------------TSPFG-------------------- 494
GQ P GFG T +Q T+PF
Sbjct: 390 RSTGQSPEGAGFGVTNSQPSIFSTSPAFSQTPVNPTNPFSQTTPTSNTNFSPSFSQPTTP 449
Query: 495 GAAQPSQPGFGSSIFGSPTAFGASS 569
QP+ P F S++ + + FG+SS
Sbjct: 450 SFGQPTTPSFRSTVSNTTSVFGSSS 474
Score = 49.3 bits (116), Expect = 3e-05
Identities = 41/157 (26%), Positives = 62/157 (39%), Gaps = 4/157 (2%)
Frame = +3
Query: 93 MFGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTG 272
+FGS Q ++ F + +FGQN + + + FG NT P Q+ T T G
Sbjct: 514 LFGSNPSFAQNTTPAFSQTSPLFGQNTTPALGQSSSVFGQNTNPALVQSNTFSTPSTGFG 573
Query: 273 VLGAAQQSSPFSSTTAFGASSSPAFGSSAPAL----GSTSTPTFGNSSSSSFGGSSVFGQ 440
+ FSS+++ S SP FG PA+ + T G ++FG + +
Sbjct: 574 --------NTFSSSSSLTTSISP-FGQITPAVTPFQSAQPTQPLGAFGFNNFGQTQIANT 624
Query: 441 KPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPT 551
G G+ F QP G+S PT
Sbjct: 625 TDIAGAMGT-------FSQGNFKQQPALGNSAVMQPT 654
Score = 41.6 bits (96), Expect = 0.007
Identities = 53/179 (29%), Positives = 65/179 (35%), Gaps = 48/179 (26%)
Frame = +3
Query: 96 FGSTNPL-GQQSSSPFGSSQSVFGQN-----------------------NSSSNPLAPKP 203
F T+PL GQ ++ G S SVFGQN +SSS + P
Sbjct: 529 FSQTSPLFGQNTTPALGQSSSVFGQNTNPALVQSNTFSTPSTGFGNTFSSSSSLTTSISP 588
Query: 204 FGSNT---TPFGAQTGTSMCGGTSTGVLGAAQQSSPFSSTTAFGASSS------PAFGSS 356
FG T TPF + T G G Q ++ A G S PA G+S
Sbjct: 589 FGQITPAVTPFQSAQPTQPLGAFGFNNFGQTQIANTTDIAGAMGTFSQGNFKQQPALGNS 648
Query: 357 A--------------PALGSTSTPTFGNSSSSSFGGSSV-FGQKPAWGGFGSTPTQTSP 488
A PAL S GNS S +G SS+ KPA P + SP
Sbjct: 649 AVMQPTPVTNPFGTLPALPQISIAQGGNSPSIQYGISSMPVVDKPA-------PVRVSP 700
Score = 41.2 bits (95), Expect = 0.009
Identities = 47/158 (29%), Positives = 68/158 (42%), Gaps = 7/158 (4%)
Frame = +3
Query: 117 GQQSS--SPFGSSQSVFGQNNSSSNPLAPKPFGSNT-----TPFGAQTGTSMCGGTSTGV 275
GQ+S+ SP G+ FG NS + + P S T PF T TS +T
Sbjct: 388 GQRSTGQSPEGAG---FGVTNSQPSIFSTSPAFSQTPVNPTNPFSQTTPTS-----NTNF 439
Query: 276 LGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQKPAWG 455
+ Q + T +FG ++P+F S+ S +T FG+SSS + S G
Sbjct: 440 SPSFSQPT----TPSFGQPTTPSFRSTV----SNTTSVFGSSSSLTTNTSQPLGSSI--- 488
Query: 456 GFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
FGSTP S G + S +FGS +F ++
Sbjct: 489 -FGSTPAHGSTPGFSIGGFNNSQSSPLFGSNPSFAQNT 525
>ref|NP_172510.2| unknown protein; protein id: At1g10390.1, supported by cDNA:
gi_19310422 [Arabidopsis thaliana]
gi|19310423|gb|AAL84948.1| At1g10390/F14N23_29
[Arabidopsis thaliana] gi|27764948|gb|AAO23595.1|
At1g10390/F14N23_29 [Arabidopsis thaliana]
Length = 1041
Score = 162 bits (411), Expect = 2e-39
Identities = 98/163 (60%), Positives = 119/163 (72%), Gaps = 4/163 (2%)
Frame = +3
Query: 93 MFGSTNPLGQQS-SSPFGSSQSVFGQ--NNSSSNPLAPK-PFGSNTTPFGAQTGTSMCGG 260
MFGS+NP GQ S +SPFGS QS+FGQ N SS+NP AP PFG++ PF AQ+G+S+ G
Sbjct: 1 MFGSSNPFGQSSGTSPFGS-QSLFGQTSNTSSNNPFAPATPFGTSA-PFAAQSGSSIFGS 58
Query: 261 TSTGVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQ 440
TSTGV GA Q SSPF+ST FGASSSPAFG+S PA G+ + +SS FGGSS FGQ
Sbjct: 59 TSTGVFGAPQTSSPFASTPTFGASSSPAFGNSTPAFGA-------SPASSPFGGSSGFGQ 111
Query: 441 KPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
KP GF STP Q++PFG + Q SQP FG++ FGS T FGA++
Sbjct: 112 KPL--GF-STP-QSNPFGNSTQQSQPAFGNTSFGSSTPFGATN 150
Score = 100 bits (250), Expect = 1e-20
Identities = 80/181 (44%), Positives = 95/181 (52%), Gaps = 28/181 (15%)
Frame = +3
Query: 102 STNPLGQQSSSPFGSSQS-VFGQNNSSSNPLAPKP-FGSNTTPFGAQTGTSMCGGTSTGV 275
ST G S+ FG++ + FG +NS S P FG++ TP TGT+ G T G
Sbjct: 166 STPSFGASSTPAFGATNTPAFGASNSPSFGATNTPAFGASPTPAFGSTGTTF-GNTGFGS 224
Query: 276 LGA--AQQSSPF--SSTTAFGASSSPAFG-SSAPALGSTSTPTFGNSSSSSFGGSSV--- 431
GA A + F S T AFGAS +PAFG SS PA G++STP FG SS+ +FGGSS
Sbjct: 225 GGAFGASNTPAFGASGTPAFGASGTPAFGASSTPAFGASSTPAFGASSTPAFGGSSTPSF 284
Query: 432 ---------FGQKPAWG---------GFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAF 557
FG PA+G FGSTP SPFGG AQ S P FG S FG T
Sbjct: 285 GASNTSSFSFGSSPAFGQSTSAFGSSAFGSTP---SPFGG-AQASTPTFGGSGFGQSTFG 340
Query: 558 G 560
G
Sbjct: 341 G 341
Score = 87.8 bits (216), Expect = 9e-17
Identities = 69/180 (38%), Positives = 91/180 (50%), Gaps = 22/180 (12%)
Frame = +3
Query: 96 FGSTNPL--GQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNT---------------TP 224
FG++ P +SSPFG S S FGQ + PFG++T TP
Sbjct: 87 FGNSTPAFGASPASSPFGGS-SGFGQKPLGFSTPQSNPFGNSTQQSQPAFGNTSFGSSTP 145
Query: 225 FGAQTGTSMCGGTSTGVLGAAQQSS-PFSSTTAFGASSSPAFG-SSAPALGSTSTPTFGN 398
FGA T T G ST GA S SST AFGA+++PAFG S++P+ G+T+TP FG
Sbjct: 146 FGA-TNTPAFGAPSTPSFGATSTPSFGASSTPAFGATNTPAFGASNSPSFGATNTPAFGA 204
Query: 399 SSSSSFGGS-SVFGQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFG--SPTAFGASS 569
S + +FG + + FG G + T FG + P+ G+ FG S AFGASS
Sbjct: 205 SPTPAFGSTGTTFGNTGFGSGGAFGASNTPAFGASGTPAFGASGTPAFGASSTPAFGASS 264
Score = 74.3 bits (181), Expect = 1e-12
Identities = 73/177 (41%), Positives = 88/177 (49%), Gaps = 34/177 (19%)
Frame = +3
Query: 132 SPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGT--SMCGGTSTGVLGAAQQSSPF 305
+PF S + FGQ +S+NP P ++T PF QT T S GT+T G SSPF
Sbjct: 419 NPFSPSPA-FGQ--TSANPTNPFSSSTSTNPFAPQTPTIASSSFGTATSNFG----SSPF 471
Query: 306 ------------SSTTAFGASSSPAFGSSAPA--LGSTSTPTFGNSSSSSFGGSSVFGQK 443
SSTT SS AFG++ P+ GS+STP FG SSSS FG + G
Sbjct: 472 GVTSSSNLFGSGSSTTTSVFGSSSAFGTTTPSPLFGSSSTPGFG-SSSSIFGSAPGQGAT 530
Query: 444 PAWGG------FGSTPT--QT--------SPFGGAAQPSQPGFG-SSIFGSP-TAFG 560
PA+G F STP+ QT S FG Q S P FG +SIF P T FG
Sbjct: 531 PAFGNSQPSTLFNSTPSTGQTGSAFGQTGSAFGQFGQSSAPAFGQNSIFNKPSTGFG 587
Score = 68.6 bits (166), Expect = 5e-11
Identities = 62/191 (32%), Positives = 86/191 (44%), Gaps = 33/191 (17%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKP-------FGSNTT-----PFGAQT 239
F + GQ S++P S ++S+NP AP+ FG+ T+ PFG +
Sbjct: 421 FSPSPAFGQTSANPTNPFSS-----STSTNPFAPQTPTIASSSFGTATSNFGSSPFGVTS 475
Query: 240 GTSMCGGTSTGVLGAAQQSSPFSSTTA---FGASSSPAFGSSAPALGST----STPTFGN 398
+++ G S+ SS F +TT FG+SS+P FGSS+ GS +TP FGN
Sbjct: 476 SSNLFGSGSSTTTSVFGSSSAFGTTTPSPLFGSSSTPGFGSSSSIFGSAPGQGATPAFGN 535
Query: 399 SSSSSFGGSSVFGQKPAWGGFGSTPTQT-SPFGGAAQPSQP-------------GFGSSI 536
S S+ +F P+ G GS QT S FG Q S P GFG+
Sbjct: 536 SQPST-----LFNSTPSTGQTGSAFGQTGSAFGQFGQSSAPAFGQNSIFNKPSTGFGNMF 590
Query: 537 FGSPTAFGASS 569
S T +SS
Sbjct: 591 SSSSTLTTSSS 601
Score = 59.7 bits (143), Expect = 3e-08
Identities = 57/165 (34%), Positives = 83/165 (49%), Gaps = 15/165 (9%)
Frame = +3
Query: 93 MFGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTG 272
+FGS++ G + SP +FG SSS P FGS+++ FG+ G +T
Sbjct: 490 VFGSSSAFGTTTPSP------LFG---SSSTP----GFGSSSSIFGSAPGQG-----ATP 531
Query: 273 VLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQKPAW 452
G +Q S+ F+ST + G + S AFG + A G FG SS+ +FG +S+F KP+
Sbjct: 532 AFGNSQPSTLFNSTPSTGQTGS-AFGQTGSAFGQ-----FGQSSAPAFGQNSIF-NKPS- 583
Query: 453 GGFGS--------TPTQTSPFGGAA-------QPSQPGFGSSIFG 542
GFG+ T + +SPFG Q +QPG S+ FG
Sbjct: 584 TGFGNMFSSSSTLTTSSSSPFGQTMPAGVTPFQSAQPGQASNGFG 628
Score = 51.6 bits (122), Expect = 7e-06
Identities = 61/226 (26%), Positives = 84/226 (36%), Gaps = 68/226 (30%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQS-VFGQNNSSSNPLAPKP-FGSNTTPFGAQTGTSMCGGTST 269
FGS G ++ FG+S + FG + + + + P FG+++TP + T GG+ST
Sbjct: 222 FGSGGAFGASNTPAFGASGTPAFGASGTPAFGASSTPAFGASSTPAFGASSTPAFGGSST 281
Query: 270 GVLGAAQQSS-PFSSTTAFGASSSP----AFGSSAPALGST--STP----------TFGN 398
GA+ SS F S+ AFG S+S AFGS+ G STP TFG
Sbjct: 282 PSFGASNTSSFSFGSSPAFGQSTSAFGSSAFGSTPSPFGGAQASTPTFGGSGFGQSTFGG 341
Query: 399 SSSSSF-------------------------------------------------GGSSV 431
S GG
Sbjct: 342 QQGGSRAVPYAPTVEADTGTGTQPAGKLESISAMPAYKEKNYEELRWEDYQRGDKGGPLP 401
Query: 432 FGQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
GQ P GFG +P+Q +PF PS P FG + F +S+
Sbjct: 402 AGQSPGNAGFGISPSQPNPF----SPS-PAFGQTSANPTNPFSSST 442
Score = 43.5 bits (101), Expect = 0.002
Identities = 55/188 (29%), Positives = 71/188 (37%), Gaps = 55/188 (29%)
Frame = +3
Query: 96 FGSTNP---LGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGS-------NTTPFGAQTGT 245
FG+T P G S+ FGSS S+FG S+ A FG+ N+TP QTG+
Sbjct: 497 FGTTTPSPLFGSSSTPGFGSSSSIFG---SAPGQGATPAFGNSQPSTLFNSTPSTGQTGS 553
Query: 246 SMCGGTSTGVLGAAQQSSP------------------FSSTTAFGASSSPAFGSSAPALG 371
+ G T + Q S+P FSS++ SSS FG + PA G
Sbjct: 554 AF-GQTGSAFGQFGQSSAPAFGQNSIFNKPSTGFGNMFSSSSTLTTSSSSPFGQTMPA-G 611
Query: 372 ST-----------------------STPTFGNSSSSSFGGSSVFGQKPAWGGFGSTPTQ- 479
T + T GN+ G FGQ PA S Q
Sbjct: 612 VTPFQSAQPGQASNGFGFNNFGQNQAANTTGNAGGLGIFGQGNFGQSPA--PLNSVVLQP 669
Query: 480 ---TSPFG 494
T+PFG
Sbjct: 670 VAVTNPFG 677
Score = 39.7 bits (91), Expect = 0.027
Identities = 45/147 (30%), Positives = 60/147 (40%)
Frame = +3
Query: 93 MFGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTG 272
+F ST GQ S+ FG + S FGQ FG ++ P Q S+ STG
Sbjct: 541 LFNSTPSTGQTGSA-FGQTGSAFGQ------------FGQSSAPAFGQ--NSIFNKPSTG 585
Query: 273 VLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQKPAW 452
+ FSS++ SSS FG + PA G T + +S+ G + FGQ A
Sbjct: 586 F------GNMFSSSSTLTTSSSSPFGQTMPA-GVTPFQSAQPGQASNGFGFNNFGQNQAA 638
Query: 453 GGFGSTPTQTSPFGGAAQPSQPGFGSS 533
G+ GG Q FG S
Sbjct: 639 NTTGNA-------GGLGIFGQGNFGQS 658
>pir||C96620 protein T30E16.23 [imported] - Arabidopsis thaliana
gi|8778749|gb|AAF79757.1|AC009317_16 T30E16.23
[Arabidopsis thaliana]
Length = 1076
Score = 155 bits (392), Expect = 3e-37
Identities = 92/155 (59%), Positives = 110/155 (70%), Gaps = 4/155 (2%)
Frame = +3
Query: 117 GQQS-SSPFGS-SQSVFGQ--NNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTGVLGA 284
GQ S SSPFG+ + S+FGQ NN+S+NP A KPFG++T PFGAQTG+SM GGTSTGV GA
Sbjct: 25 GQSSISSPFGTQTHSLFGQTNNNASNNPFATKPFGTST-PFGAQTGSSMFGGTSTGVFGA 83
Query: 285 AQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQKPAWGGFG 464
Q SSPF +S AFGSS A G++STP+FG SS+S FGG+S FGQK FG
Sbjct: 84 PQTSSPF-------GASPQAFGSSTQAFGASSTPSFG-SSNSPFGGTSTFGQK----SFG 131
Query: 465 STPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
+ Q+SPFG Q SQP FG+S FGS T FGAS+
Sbjct: 132 LSTPQSSPFGSTTQQSQPAFGNSTFGSSTPFGAST 166
Score = 90.5 bits (223), Expect = 1e-17
Identities = 70/167 (41%), Positives = 97/167 (57%), Gaps = 9/167 (5%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQS-VFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTG 272
FGS+ P G ++ FG+S + FG +N+S FG+ TP T T+ GG+ST
Sbjct: 156 FGSSTPFGASTTPAFGASSTPAFGVSNTSG-------FGATNTPGFGATNTTGFGGSSTP 208
Query: 273 VLGAAQQSSPFSSTT-AFGASSSPAFG-SSAPALGSTSTPTFGNSSSSSFGGSSVFGQKP 446
GA+ + S+ T AFGASS+P FG SS+PA G++ P FG+S ++ G++ F
Sbjct: 209 GFGASSTPAFGSTNTPAFGASSTPLFGSSSSPAFGASPAPAFGSSGNAF--GNNTFSSGG 266
Query: 447 AWGGFGSTPT----QTSPFGGAAQPSQPGFGSS-IFG-SPTAFGASS 569
A+G STPT TS FG ++ PS FGSS FG S +AFG+SS
Sbjct: 267 AFGS-SSTPTFGASNTSAFGASSSPSF-NFGSSPAFGQSTSAFGSSS 311
Score = 65.1 bits (157), Expect = 6e-10
Identities = 49/124 (39%), Positives = 69/124 (55%), Gaps = 5/124 (4%)
Frame = +3
Query: 102 STNPLGQQSSSPFGSSQS-VFGQNNSSSNPLAPKP-FGSNTTPFGAQTGTS--MCGGTST 269
ST G ++ FG+S + +FG ++S + +P P FGS+ FG T +S G +ST
Sbjct: 214 STPAFGSTNTPAFGASSTPLFGSSSSPAFGASPAPAFGSSGNAFGNNTFSSGGAFGSSST 273
Query: 270 GVLGAAQQSSPFSSTTAFGASSSPAFG-SSAPALGSTSTPTFGNSSSSSFGGSSVFGQKP 446
GA S+T+AFGASSSP+F S+PA G ST FG+SS + +S FG +
Sbjct: 274 PTFGA-------SNTSAFGASSSPSFNFGSSPAFGQ-STSAFGSSSFGAQASTSTFGGQS 325
Query: 447 AWGG 458
GG
Sbjct: 326 TIGG 329
Score = 60.1 bits (144), Expect = 2e-08
Identities = 58/181 (32%), Positives = 81/181 (44%), Gaps = 34/181 (18%)
Frame = +3
Query: 102 STNPLGQQSSSPFGSS----QSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTST 269
+T GQ ++ F S+ SVFG ++S + + +P GS+ FG+ G S
Sbjct: 477 TTPSFGQPTTPSFRSTVSNTTSVFGSSSSLTTNTS-QPLGSSI--FGSTPAHGSTPGFSI 533
Query: 270 GVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQ--K 443
G +Q S F S +F +++PAF ++P G +TP G S SSVFGQ
Sbjct: 534 GGFNNSQSSPLFGSNPSFAQNTTPAFSQTSPLFGQNTTPALGQS-------SSVFGQNTN 586
Query: 444 PAW----------GGFGST-------PTQTSPFG---------GAAQPSQP--GFGSSIF 539
PA GFG+T T SPFG +AQP+QP FG + F
Sbjct: 587 PALVQSNTFSTPSTGFGNTFSSSSSLTTSISPFGQITPAVTPFQSAQPTQPLGAFGFNNF 646
Query: 540 G 542
G
Sbjct: 647 G 647
Score = 55.5 bits (132), Expect = 5e-07
Identities = 53/152 (34%), Positives = 72/152 (46%), Gaps = 10/152 (6%)
Frame = +3
Query: 96 FGSTNP--LGQQSSSPFGSSQS-VFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMC---G 257
FGSTN G S+ FGSS S FG + + + + FG+NT G G+S G
Sbjct: 218 FGSTNTPAFGASSTPLFGSSSSPAFGASPAPAFGSSGNAFGNNTFSSGGAFGSSSTPTFG 277
Query: 258 GTSTGVLGAAQQSS-PFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSS--SSFGGSS 428
++T GA+ S F S+ AFG S+S AFGSS+ ST TFG S+ GGS
Sbjct: 278 ASNTSAFGASSSPSFNFGSSPAFGQSTS-AFGSSSFG-AQASTSTFGGQSTIGGQQGGSR 335
Query: 429 VFGQKPAWGGFGSTPTQTSPFGG-AAQPSQPG 521
V P T +++ +A P+ G
Sbjct: 336 VIPYAPTTDTASGTESKSERLQSISAMPAHKG 367
Score = 50.4 bits (119), Expect = 2e-05
Identities = 57/170 (33%), Positives = 75/170 (43%), Gaps = 18/170 (10%)
Frame = +3
Query: 96 FGSTNPLGQQ-SSSPFGSSQSV-----FGQNNSSSNPLAPKPFGSNTTP-FGAQTGTSMC 254
FG TN S+SP S V F Q +SN F TTP FG T S
Sbjct: 431 FGVTNSQPSIFSTSPAFSQTPVNPTNPFSQTTPTSNTNFSPSFSQPTTPSFGQPTTPSF- 489
Query: 255 GGTSTGVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGST---STPTFGNSSSSSFGG- 422
+S+ ++T+ FG+SSS +S P LGS+ STP G++ S GG
Sbjct: 490 ------------RSTVSNTTSVFGSSSSLTTNTSQP-LGSSIFGSTPAHGSTPGFSIGGF 536
Query: 423 -----SSVFGQKPAWGGFGSTP--TQTSPFGGAAQPSQPGFGSSIFGSPT 551
S +FG P++ +TP +QTSP G G SS+FG T
Sbjct: 537 NNSQSSPLFGSNPSFAQ-NTTPAFSQTSPLFGQNTTPALGQSSSVFGQNT 585
Score = 43.9 bits (102), Expect = 0.001
Identities = 32/117 (27%), Positives = 51/117 (43%), Gaps = 4/117 (3%)
Frame = +3
Query: 93 MFGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTG 272
+FGS Q ++ F + +FGQN + + + FG NT P Q+ T T G
Sbjct: 544 LFGSNPSFAQNTTPAFSQTSPLFGQNTTPALGQSSSVFGQNTNPALVQSNTFSTPSTGFG 603
Query: 273 VLGAAQQSSPFSSTTAFGASSSPAFGSSAPAL----GSTSTPTFGNSSSSSFGGSSV 431
+ FSS+++ S SP FG PA+ + T G ++FG + +
Sbjct: 604 --------NTFSSSSSLTTSISP-FGQITPAVTPFQSAQPTQPLGAFGFNNFGQTQI 651
Score = 41.2 bits (95), Expect = 0.009
Identities = 47/158 (29%), Positives = 68/158 (42%), Gaps = 7/158 (4%)
Frame = +3
Query: 117 GQQSS--SPFGSSQSVFGQNNSSSNPLAPKPFGSNT-----TPFGAQTGTSMCGGTSTGV 275
GQ+S+ SP G+ FG NS + + P S T PF T TS +T
Sbjct: 418 GQRSTGQSPEGAG---FGVTNSQPSIFSTSPAFSQTPVNPTNPFSQTTPTS-----NTNF 469
Query: 276 LGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVFGQKPAWG 455
+ Q + T +FG ++P+F S+ S +T FG+SSS + S G
Sbjct: 470 SPSFSQPT----TPSFGQPTTPSFRSTV----SNTTSVFGSSSSLTTNTSQPLGSSI--- 518
Query: 456 GFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
FGSTP S G + S +FGS +F ++
Sbjct: 519 -FGSTPAHGSTPGFSIGGFNNSQSSPLFGSNPSFAQNT 555
Score = 31.2 bits (69), Expect = 9.7
Identities = 30/108 (27%), Positives = 45/108 (40%), Gaps = 11/108 (10%)
Frame = +3
Query: 279 GAAQQSSPFSSTTAFGASSS-PAFGSSAPALGST----------STPTFGNSSSSSFGGS 425
G + + FG ++S P+ S++PA T +TPT + S SF
Sbjct: 417 GGQRSTGQSPEGAGFGVTNSQPSIFSTSPAFSQTPVNPTNPFSQTTPTSNTNFSPSF--- 473
Query: 426 SVFGQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
S PT T FG QP+ P F S++ + + FG+SS
Sbjct: 474 -------------SQPT-TPSFG---QPTTPSFRSTVSNTTSVFGSSS 504
>pir||H86237 protein F14N23.29 [imported] - Arabidopsis thaliana
gi|4914343|gb|AAD32891.1|AC005489_29 F14N23.29
[Arabidopsis thaliana]
Length = 1096
Score = 139 bits (350), Expect = 3e-32
Identities = 93/176 (52%), Positives = 114/176 (63%), Gaps = 25/176 (14%)
Frame = +3
Query: 117 GQQS-SSPFGSSQSVFGQ--NNSSSNPLAPK-PFGSNTTPFGAQTGTSMCGGTSTGVLGA 284
GQ S +SPFGS QS+FGQ N SS+NP AP PFG++ PF AQ+G+S+ G TSTGV GA
Sbjct: 27 GQSSGTSPFGS-QSLFGQTSNTSSNNPFAPATPFGTSA-PFAAQSGSSIFGSTSTGVFGA 84
Query: 285 AQQSSPFSSTTAFGASSSPAFGSSAPALGST--STPTFGNSSSS---------------- 410
Q SSPF+ST FGASSSPAFG+S PA G++ S+P G +
Sbjct: 85 PQTSSPFASTPTFGASSSPAFGNSTPAFGASPASSPFGGQGFKNKPCLTVLHLHLVIKLI 144
Query: 411 ---SFGGSSVFGQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
SF GSS FGQKP GF STP Q++PFG + Q SQP FG++ FGS T FGA++
Sbjct: 145 LFVSFLGSSGFGQKPL--GF-STP-QSNPFGNSTQQSQPAFGNTSFGSSTPFGATN 196
Score = 85.1 bits (209), Expect = 6e-16
Identities = 63/163 (38%), Positives = 85/163 (51%), Gaps = 10/163 (6%)
Frame = +3
Query: 111 PLGQQS--SSPFGSS----QSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSMCGGTSTG 272
PLG + S+PFG+S Q FG + S+ PFG+ TP T G TST
Sbjct: 159 PLGFSTPQSNPFGNSTQQSQPAFGNTSFGSST----PFGATNTPAFGAPSTPSFGATSTP 214
Query: 273 VLGAAQQSSPFSSTTAFGASSSPAFG-SSAPALGSTSTPTFGNSSSSSFGGS-SVFGQKP 446
GA SST AFGA+++PAFG S++P+ G+T+TP FG S + +FG + + FG
Sbjct: 215 SFGA-------SSTPAFGATNTPAFGASNSPSFGATNTPAFGASPTPAFGSTGTTFGNTG 267
Query: 447 AWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFG--SPTAFGASS 569
G + T FG + P+ G+ FG S AFGASS
Sbjct: 268 FGSGGAFGASNTPAFGASGTPAFGASGTPAFGASSTPAFGASS 310
Score = 80.5 bits (197), Expect = 1e-14
Identities = 66/193 (34%), Positives = 90/193 (46%), Gaps = 35/193 (18%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPF----------------------- 206
F ST G SS FG+S FG + +SS P + F
Sbjct: 91 FASTPTFGASSSPAFGNSTPAFGASPASS-PFGGQGFKNKPCLTVLHLHLVIKLILFVSF 149
Query: 207 ----GSNTTPFGAQTGTSMCGGTSTGVLGAAQQSSPFSSTTAFGASSSPAFGS-SAPALG 371
G P G T S G ST A ++ F S+T FGA+++PAFG+ S P+ G
Sbjct: 150 LGSSGFGQKPLGFSTPQSNPFGNSTQQSQPAFGNTSFGSSTPFGATNTPAFGAPSTPSFG 209
Query: 372 STSTPTFGNSSSSSFGGSSVFGQKPAWGGFGST---PTQTSPFGGAAQP----SQPGFGS 530
+TSTP+FG SS+ +FG ++ PA+G S T T FG + P + FG+
Sbjct: 210 ATSTPSFGASSTPAFGATNT----PAFGASNSPSFGATNTPAFGASPTPAFGSTGTTFGN 265
Query: 531 SIFGSPTAFGASS 569
+ FGS AFGAS+
Sbjct: 266 TGFGSGGAFGASN 278
Score = 76.6 bits (187), Expect = 2e-13
Identities = 64/172 (37%), Positives = 80/172 (46%), Gaps = 17/172 (9%)
Frame = +3
Query: 96 FGSTNP--LGQQSSSPFGSSQSVFGQNNSSSNPLA----PKPFGSNTTPFGAQTGTSMCG 257
FG+TN G + FGS+ + FG S FG++ TP +GT G
Sbjct: 240 FGATNTPAFGASPTPAFGSTGTTFGNTGFGSGGAFGASNTPAFGASGTPAFGASGTPAFG 299
Query: 258 GTSTGVLGAAQQSS-PFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFGGSSVF 434
+ST GA+ + SST AFG SS+P+FG+S N+SS SFG S F
Sbjct: 300 ASSTPAFGASSTPAFGASSTPAFGGSSTPSFGAS-------------NTSSFSFGSSPAF 346
Query: 435 GQKPAWGG---FGSTPTQTSPFGG-------AAQPSQPGFGSSIFGSPTAFG 560
GQ + G FGSTP SPFGG + S P FG S FG T G
Sbjct: 347 GQSTSAFGSSAFGSTP---SPFGGQGSSFSISHSASTPTFGGSGFGQSTFGG 395
Score = 74.3 bits (181), Expect = 1e-12
Identities = 73/177 (41%), Positives = 88/177 (49%), Gaps = 34/177 (19%)
Frame = +3
Query: 132 SPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGT--SMCGGTSTGVLGAAQQSSPF 305
+PF S + FGQ +S+NP P ++T PF QT T S GT+T G SSPF
Sbjct: 473 NPFSPSPA-FGQ--TSANPTNPFSSSTSTNPFAPQTPTIASSSFGTATSNFG----SSPF 525
Query: 306 ------------SSTTAFGASSSPAFGSSAPA--LGSTSTPTFGNSSSSSFGGSSVFGQK 443
SSTT SS AFG++ P+ GS+STP FG SSSS FG + G
Sbjct: 526 GVTSSSNLFGSGSSTTTSVFGSSSAFGTTTPSPLFGSSSTPGFG-SSSSIFGSAPGQGAT 584
Query: 444 PAWGG------FGSTPT--QT--------SPFGGAAQPSQPGFG-SSIFGSP-TAFG 560
PA+G F STP+ QT S FG Q S P FG +SIF P T FG
Sbjct: 585 PAFGNSQPSTLFNSTPSTGQTGSAFGQTGSAFGQFGQSSAPAFGQNSIFNKPSTGFG 641
Score = 68.6 bits (166), Expect = 5e-11
Identities = 62/191 (32%), Positives = 86/191 (44%), Gaps = 33/191 (17%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKP-------FGSNTT-----PFGAQT 239
F + GQ S++P S ++S+NP AP+ FG+ T+ PFG +
Sbjct: 475 FSPSPAFGQTSANPTNPFSS-----STSTNPFAPQTPTIASSSFGTATSNFGSSPFGVTS 529
Query: 240 GTSMCGGTSTGVLGAAQQSSPFSSTTA---FGASSSPAFGSSAPALGST----STPTFGN 398
+++ G S+ SS F +TT FG+SS+P FGSS+ GS +TP FGN
Sbjct: 530 SSNLFGSGSSTTTSVFGSSSAFGTTTPSPLFGSSSTPGFGSSSSIFGSAPGQGATPAFGN 589
Query: 399 SSSSSFGGSSVFGQKPAWGGFGSTPTQT-SPFGGAAQPSQP-------------GFGSSI 536
S S+ +F P+ G GS QT S FG Q S P GFG+
Sbjct: 590 SQPST-----LFNSTPSTGQTGSAFGQTGSAFGQFGQSSAPAFGQNSIFNKPSTGFGNMF 644
Query: 537 FGSPTAFGASS 569
S T +SS
Sbjct: 645 SSSSTLTTSSS 655
Score = 58.2 bits (139), Expect = 7e-08
Identities = 63/170 (37%), Positives = 82/170 (48%), Gaps = 16/170 (9%)
Frame = +3
Query: 93 MFGSTNPLGQQSSSP-FGSSQSV-FGQNNSSSNPLAPKPFGSNTTPFG-AQTGTSMCGGT 263
+FGS++ G + SP FGSS + FG SSS+ P T FG +Q T
Sbjct: 544 VFGSSSAFGTTTPSPLFGSSSTPGFG---SSSSIFGSAPGQGATPAFGNSQPSTLFNSTP 600
Query: 264 STGVLGAA--QQSSPFSSTTAFGASSSPAFGSSA----PALG----STSTPTFGNSSSSS 413
STG G+A Q S F FG SS+PAFG ++ P+ G +S+ T SSSS
Sbjct: 601 STGQTGSAFGQTGSAFGQ---FGQSSAPAFGQNSIFNKPSTGFGNMFSSSSTLTTSSSSP 657
Query: 414 FGGSSV---FGQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTA 554
FG + V F KP G TP Q++ G A+ GFG + FG A
Sbjct: 658 FGQTMVIFTFAIKPKHFPAGVTPFQSAQPGQASN----GFGFNNFGQNQA 703
Score = 50.1 bits (118), Expect = 2e-05
Identities = 62/234 (26%), Positives = 85/234 (35%), Gaps = 76/234 (32%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQS-VFGQNNSSSNPLAPKP-FGSNTTPFGAQTGTSMCGGTST 269
FGS G ++ FG+S + FG + + + + P FG+++TP + T GG+ST
Sbjct: 268 FGSGGAFGASNTPAFGASGTPAFGASGTPAFGASSTPAFGASSTPAFGASSTPAFGGSST 327
Query: 270 GVLGAAQQSS-PFSSTTAFGASSSP----AFGSSAPALG----------STSTP------ 386
GA+ SS F S+ AFG S+S AFGS+ G S STP
Sbjct: 328 PSFGASNTSSFSFGSSPAFGQSTSAFGSSAFGSTPSPFGGQGSSFSISHSASTPTFGGSG 387
Query: 387 ----TFGNSSSSSF---------------------------------------------- 416
TFG S
Sbjct: 388 FGQSTFGGQQGGSRAVPYAPTVEADTGTGTQPAGKLESISAMPAYKEKNYEELRWEDYQR 447
Query: 417 ---GGSSVFGQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGASS 569
GG GQ P GFG +P+Q +PF PS P FG + F +S+
Sbjct: 448 GDKGGPLPAGQSPGNAGFGISPSQPNPF----SPS-PAFGQTSANPTNPFSSST 496
Score = 40.0 bits (92), Expect = 0.021
Identities = 39/134 (29%), Positives = 56/134 (41%), Gaps = 21/134 (15%)
Frame = +3
Query: 93 MFGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPF-GAQTGTSMCG---- 257
MF S++ L SSSPFG + +F + + PK F + TPF AQ G + G
Sbjct: 643 MFSSSSTLTTSSSSPFGQTMVIF------TFAIKPKHFPAGVTPFQSAQPGQASNGFGFN 696
Query: 258 -------GTSTGVLGA---------AQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPT 389
+TG G Q +P +S + + FG + PA+ S
Sbjct: 697 NFGQNQAANTTGNAGGLGIFGQGNFGQSPAPLNSVVLQPVAVTNPFG-TLPAMPQISINQ 755
Query: 390 FGNSSSSSFGGSSV 431
GNS S +G SS+
Sbjct: 756 GGNSPSIQYGISSM 769
>emb|CAB53357.1| nucleoporin CAN [Xenopus laevis]
Length = 2037
Score = 73.9 bits (180), Expect = 1e-12
Identities = 63/174 (36%), Positives = 75/174 (42%), Gaps = 22/174 (12%)
Frame = +3
Query: 108 NPLGQQSSSPFGSSQSVFGQNNSSS--NPLAPKPFGSNTTPFG-------------AQTG 242
NP G SS FGS+ G +NSS+ K FG T FG A G
Sbjct: 1843 NPFGSPGSSGFGSA----GASNSSNLFGNSGAKAFGFGGTSFGDKPSATFSAGGSVASQG 1898
Query: 243 TSMCGGTSTGVLGAAQQSSPFSSTTAFGAS----SSPAFGSSAP---ALGSTSTPTFGNS 401
S T TG GAA F S FG S SPAFG++A LGST FG
Sbjct: 1899 FSFNSPTKTGGFGAAPV---FGSPPTFGGSPGFGGSPAFGTAAAFSNTLGSTGGKVFGEG 1955
Query: 402 SSSSFGGSSVFGQKPAWGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGA 563
+S++ G FG + FGS TQ +P G+ PGFG G + FGA
Sbjct: 1956 TSAATTGGFGFGSNSSTAAFGSLATQNTPTFGSISQQSPGFGGQSSGF-SGFGA 2008
Score = 65.1 bits (157), Expect = 6e-10
Identities = 51/155 (32%), Positives = 78/155 (49%), Gaps = 5/155 (3%)
Frame = +3
Query: 117 GQQSSSPFGSSQSVFGQNNSSSNPL-APKPFGSNTTPFGAQTGTSMCGGTSTGVLGAAQQ 293
GQ + G+S S+F + S S + +P S+ FGA +G + G +G+ A+
Sbjct: 1778 GQPAGFASGTSGSLFNPSQSGSTSVFGQQPASSSGGLFGAGSGGASTVGLFSGL--GAKP 1835
Query: 294 SSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSFG-GSSVFGQKPA--WGGFG 464
S ++ FG+ S FGS+ S S+ FGNS + +FG G + FG KP+ + G
Sbjct: 1836 SQEAANKNPFGSPGSSGFGSAG---ASNSSNLFGNSGAKAFGFGGTSFGDKPSATFSAGG 1892
Query: 465 STPTQTSPFGGAAQPSQPGFGSS-IFGSPTAFGAS 566
S +Q F + GFG++ +FGSP FG S
Sbjct: 1893 SVASQGFSFNSPTKTG--GFGAAPVFGSPPTFGGS 1925
Score = 59.7 bits (143), Expect = 3e-08
Identities = 55/182 (30%), Positives = 76/182 (41%), Gaps = 29/182 (15%)
Frame = +3
Query: 111 PLGQQSSSPFGSSQSVFGQNNSSSNPLAPKPFGSNTTPFGAQTGTSM--------CGGTS 266
P QS+ PF SQ FG + P A S + FG + S GTS
Sbjct: 1678 PATSQSTLPF--SQPTFGTQPAFGQPAASTATSSAGSLFGCTSSASSFSFGQASNTSGTS 1735
Query: 267 TGVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPTFGNSSSSSF---------- 416
T + Q S+P +A ++PAFGS++ + +T+T +FG + F
Sbjct: 1736 TSGVLFGQSSAPVFGQSAAFPQAAPAFGSAS--VSTTTTASFGFGQPAGFASGTSGSLFN 1793
Query: 417 ----GGSSVFGQKPA-------WGGFGSTPTQTSPFGGAAQPSQPGFGSSIFGSPTAFGA 563
G +SVFGQ+PA G G T G A+PSQ + FGSP + G
Sbjct: 1794 PSQSGSTSVFGQQPASSSGGLFGAGSGGASTVGLFSGLGAKPSQEAANKNPFGSPGSSGF 1853
Query: 564 SS 569
S
Sbjct: 1854 GS 1855
Score = 53.9 bits (128), Expect = 1e-06
Identities = 52/167 (31%), Positives = 75/167 (44%), Gaps = 23/167 (13%)
Frame = +3
Query: 96 FGSTNPLGQQSSSPFGSSQSVFGQNNSSSNPL------------APKPFGSN-----TTP 224
FG T+ G + S+ F + SV Q S ++P +P FG + +
Sbjct: 1875 FGGTS-FGDKPSATFSAGGSVASQGFSFNSPTKTGGFGAAPVFGSPPTFGGSPGFGGSPA 1933
Query: 225 FGAQTGTSMCGGTSTGVLGAAQQSSPFSSTTAFGA-SSSPAFGSSAPALGSTSTPTFGNS 401
FG S G++ G + S+ + FG+ SS+ AFGS L + +TPTFG+
Sbjct: 1934 FGTAAAFSNTLGSTGGKVFGEGTSAATTGGFGFGSNSSTAAFGS----LATQNTPTFGSI 1989
Query: 402 SSSSFGGSSVFGQKPAWGGFGSTP----TQTSPFG-GAAQPSQPGFG 527
S S G GQ + GFG+ P T FG G + P+ PGFG
Sbjct: 1990 SQQSPGFG---GQSSGFSGFGAGPGAAAGNTGGFGFGVSNPTSPGFG 2033
Score = 52.8 bits (125), Expect = 3e-06
Identities = 56/162 (34%), Positives = 76/162 (46%), Gaps = 14/162 (8%)
Frame = +3
Query: 126 SSSPFGSS-------QSVFGQNNSSSNPLAPKPFGSNTT-PFGAQT-GTSMCGGTSTGVL 278
SS+ FG+S FGQ + PL P S +T PF T GT G
Sbjct: 1646 SSTGFGTSAFGATGGNGGFGQPSFGQAPLWKGPATSQSTLPFSQPTFGTQPAFGQPAAST 1705
Query: 279 GAAQQSSPFSSTTAFGASSSPAFGSSAPALG-STSTPTFGNSSSSSFGGSSVFGQ-KPAW 452
+ S F T++ +SS +FG ++ G STS FG SS+ FG S+ F Q PA+
Sbjct: 1706 ATSSAGSLFGCTSS---ASSFSFGQASNTSGTSTSGVLFGQSSAPVFGQSAAFPQAAPAF 1762
Query: 453 GGFGSTPTQTSPFGGAAQPSQPGFGSSIFGS---PTAFGASS 569
G + T T+ F G QP+ GF S GS P+ G++S
Sbjct: 1763 GSASVSTTTTASF-GFGQPA--GFASGTSGSLFNPSQSGSTS 1801
Score = 47.0 bits (110), Expect = 2e-04
Identities = 55/159 (34%), Positives = 66/159 (40%), Gaps = 27/159 (16%)
Frame = +3
Query: 174 SSSNPLAPKPFGSNT-------------TPFGAQTGTSMCGGTSTGVLGAAQQSSPFSST 314
+S PLA P G T T AQT S+ G G AQ + +ST
Sbjct: 1558 ASQTPLASTPAGGPTSQVPVLVTTAPPVTTESAQT-VSLTGQPVAGSSAFAQSTVTAAST 1616
Query: 315 TAFG-ASSSPAFGSSAPALGSTSTPTFGNSSS----SSFG--------GSSVFGQKPAWG 455
FG A +S A S S+S T NSS+ S+FG G FGQ P W
Sbjct: 1617 PVFGQALASGAAPSPFAQPTSSSVSTSANSSTGFGTSAFGATGGNGGFGQPSFGQAPLWK 1676
Query: 456 GFGSTPTQTSPFGGAAQPSQPGFGSS-IFGSPTAFGASS 569
G +T T PF SQP FG+ FG P A A+S
Sbjct: 1677 G-PATSQSTLPF------SQPTFGTQPAFGQPAASTATS 1708
Score = 37.4 bits (85), Expect = 0.13
Identities = 37/126 (29%), Positives = 49/126 (38%), Gaps = 5/126 (3%)
Frame = +3
Query: 210 SNTTPFGAQTGTSMCGGTSTGVLGAAQQSSPFSSTTAFGASSSPAFGSSAPALGSTSTPT 389
S T PF T T + T L + P S ++ P SA + T P
Sbjct: 1547 SQTVPF---TSTVLASQTP---LASTPAGGPTSQVPVLVTTAPPVTTESAQTVSLTGQPV 1600
Query: 390 FGNS----SSSSFGGSSVFGQKPAWGGFGSTPTQ-TSPFGGAAQPSQPGFGSSIFGSPTA 554
G+S S+ + + VFGQ A G S Q TS + S GFG+S FG+
Sbjct: 1601 AGSSAFAQSTVTAASTPVFGQALASGAAPSPFAQPTSSSVSTSANSSTGFGTSAFGATGG 1660
Query: 555 FGASSQ 572
G Q
Sbjct: 1661 NGGFGQ 1666
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 558,845,568
Number of Sequences: 1393205
Number of extensions: 14443127
Number of successful extensions: 97012
Number of sequences better than 10.0: 2588
Number of HSP's better than 10.0 without gapping: 62918
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 82113
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)