Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002697A_C01 KMC002697A_c01
(610 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
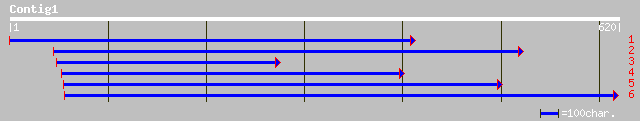
gb|AAD54273.1|AF167556_1 dihydroflavonol-4-reductase DFR1 [Glyci... 217 1e-55
emb|CAB94914.1| dihydroflavonol 4-reductase [Juglans nigra] 189 3e-47
gb|AAC25960.1| dihydroflavonol 4-reductase [Fragaria x ananassa] 181 9e-45
dbj|BAA19658.1| dihydroflavonol 4-reductase [Perilla frutescens] 180 1e-44
dbj|BAA12723.1| dihydroflavonol 4-reductase [Rosa hybrid cultivar] 177 7e-44
>gb|AAD54273.1|AF167556_1 dihydroflavonol-4-reductase DFR1 [Glycine max]
Length = 347
Score = 217 bits (552), Expect = 1e-55
Identities = 102/123 (82%), Positives = 110/123 (88%)
Frame = -2
Query: 606 TALCPITGNEAQYMIMKQSQFVHVDDLCLAHIFLFEHPESEGRYMCSACDANIHDIAKLI 427
TAL PITGNE Y I+KQ QFVH+DDLCLAHIFLFE PE EGRY+CSACDA IHDIAKLI
Sbjct: 208 TALSPITGNEDHYSIIKQGQFVHLDDLCLAHIFLFEEPEVEGRYICSACDATIHDIAKLI 267
Query: 426 NTKYPEYNVPTKFKNIPDELELVRFSSKKIKDMGFQFKYTLEDMYTGAIDACREKGLLPK 247
N KYPEY VPTKFKNIPD+LELVRFSSKKI D+GF+FKY+LEDMYTGAID CR+KGLLPK
Sbjct: 268 NQKYPEYKVPTKFKNIPDQLELVRFSSKKITDLGFKFKYSLEDMYTGAIDTCRDKGLLPK 327
Query: 246 AAE 238
AE
Sbjct: 328 PAE 330
>emb|CAB94914.1| dihydroflavonol 4-reductase [Juglans nigra]
Length = 229
Score = 189 bits (479), Expect = 3e-47
Identities = 84/123 (68%), Positives = 103/123 (83%)
Frame = -2
Query: 606 TALCPITGNEAQYMIMKQSQFVHVDDLCLAHIFLFEHPESEGRYMCSACDANIHDIAKLI 427
TAL PITGNEA Y I+KQ QFVH+DDLC+ HI+LFEHP++EGRY+CSACDA I D+AKL+
Sbjct: 104 TALSPITGNEAHYSIIKQGQFVHLDDLCMGHIYLFEHPKAEGRYLCSACDATILDVAKLL 163
Query: 426 NTKYPEYNVPTKFKNIPDELELVRFSSKKIKDMGFQFKYTLEDMYTGAIDACREKGLLPK 247
K+PE NVPTKFK + + LE++ F+SKKIKD+GFQFKY+LEDM+ A+ CR KGLLP
Sbjct: 164 REKFPECNVPTKFKGVDESLEIISFNSKKIKDLGFQFKYSLEDMFVEAVQTCRAKGLLPP 223
Query: 246 AAE 238
AAE
Sbjct: 224 AAE 226
>gb|AAC25960.1| dihydroflavonol 4-reductase [Fragaria x ananassa]
Length = 341
Score = 181 bits (458), Expect = 9e-45
Identities = 81/123 (65%), Positives = 103/123 (82%)
Frame = -2
Query: 606 TALCPITGNEAQYMIMKQSQFVHVDDLCLAHIFLFEHPESEGRYMCSACDANIHDIAKLI 427
+ L P+TGNEA Y I+KQ Q+VH+DDLC +HIFL+EH ++EGRY+CS+ DA IHDIAKL+
Sbjct: 210 SGLSPLTGNEAHYGIIKQCQYVHLDDLCQSHIFLYEHAKAEGRYICSSHDATIHDIAKLL 269
Query: 426 NTKYPEYNVPTKFKNIPDELELVRFSSKKIKDMGFQFKYTLEDMYTGAIDACREKGLLPK 247
N KYP+YNVP KFK I + L + FSSKK+K+MGF+FK++LEDM+TGA+DACREKGLLP
Sbjct: 270 NEKYPKYNVPKKFKGIEENLTNIHFSSKKLKEMGFEFKHSLEDMFTGAVDACREKGLLPL 329
Query: 246 AAE 238
E
Sbjct: 330 PQE 332
>dbj|BAA19658.1| dihydroflavonol 4-reductase [Perilla frutescens]
Length = 456
Score = 180 bits (456), Expect = 1e-44
Identities = 83/119 (69%), Positives = 100/119 (83%)
Frame = -2
Query: 606 TALCPITGNEAQYMIMKQSQFVHVDDLCLAHIFLFEHPESEGRYMCSACDANIHDIAKLI 427
TAL PITGNEA Y I+KQ QFVHVDDLC AHIFLFEHP +EGRY+CS+ DA I+DIAK+I
Sbjct: 215 TALSPITGNEAHYSIIKQGQFVHVDDLCEAHIFLFEHPAAEGRYICSSHDATIYDIAKMI 274
Query: 426 NTKYPEYNVPTKFKNIPDELELVRFSSKKIKDMGFQFKYTLEDMYTGAIDACREKGLLP 250
+PEY+VPT+F+ I ++ +V FSSKK+ DMGF FKYTLEDMY GAI++CREKG+LP
Sbjct: 275 GENWPEYHVPTEFEGIHKDIAVVSFSSKKLVDMGFSFKYTLEDMYRGAIESCREKGMLP 333
>dbj|BAA12723.1| dihydroflavonol 4-reductase [Rosa hybrid cultivar]
Length = 349
Score = 177 bits (450), Expect = 7e-44
Identities = 78/123 (63%), Positives = 100/123 (80%)
Frame = -2
Query: 606 TALCPITGNEAQYMIMKQSQFVHVDDLCLAHIFLFEHPESEGRYMCSACDANIHDIAKLI 427
T L P+TGNE+ Y I+KQ QF+H+DDLC +HI+L+EHP++EGRY+CS+ DA IH+IAKL+
Sbjct: 208 TGLSPLTGNESHYSIIKQGQFIHLDDLCQSHIYLYEHPKAEGRYICSSHDATIHEIAKLL 267
Query: 426 NTKYPEYNVPTKFKNIPDELELVRFSSKKIKDMGFQFKYTLEDMYTGAIDACREKGLLPK 247
KYPEYNVPT FK I + L V FSSKK+ + GF+FKY+LEDM+ GA+DAC+EKGLLP
Sbjct: 268 KGKYPEYNVPTTFKGIEENLPKVHFSSKKLLETGFEFKYSLEDMFVGAVDACKEKGLLPP 327
Query: 246 AAE 238
E
Sbjct: 328 PTE 330
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 535,887,343
Number of Sequences: 1393205
Number of extensions: 11831668
Number of successful extensions: 31731
Number of sequences better than 10.0: 212
Number of HSP's better than 10.0 without gapping: 30705
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31650
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24283162270
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)