Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002690A_C01 KMC002690A_c01
(874 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
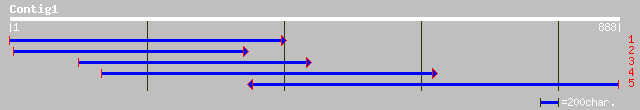
gb|AAN74837.1| Putative ubiquitin-conjugating enzyme [Oryza sati... 318 9e-86
emb|CAD29823.2| putative ubiquitin-conjugating enzyme [Populus e... 316 3e-85
gb|AAL58113.1|AC092697_1 putative ubiquitin-conjugating enzyme [... 308 7e-83
ref|NP_191346.1| E2 ubiquitin-conjugating-like enzyme, Ahus5; pr... 303 2e-81
pir||T50603 ubiquitin-conjugating enzyme [imported] - Prunus arm... 260 2e-68
>gb|AAN74837.1| Putative ubiquitin-conjugating enzyme [Oryza sativa (japonica
cultivar-group)]
Length = 160
Score = 318 bits (814), Expect = 9e-86
Identities = 142/157 (90%), Positives = 151/157 (95%)
Frame = -3
Query: 755 GIARGRLTEERKSWRKNHPHGFVAKPETLSDGTINLMVWHCTIPGKTGTDWDGGYFPLTL 576
GIARGRL EERK+WRKNHPHGFVAKPETL+DGT+NLM+WHCTIPGK GTDW+GGYFPLTL
Sbjct: 4 GIARGRLAEERKAWRKNHPHGFVAKPETLADGTVNLMIWHCTIPGKQGTDWEGGYFPLTL 63
Query: 575 HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ 396
HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ
Sbjct: 64 HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ 123
Query: 395 PNPADPAQTEGYHLFIQDVAEYKRRVQQQAKLFPPLV 285
PNPADPAQT+GYHLFIQD EYKRRV+ QAK +PP+V
Sbjct: 124 PNPADPAQTDGYHLFIQDPTEYKRRVRLQAKQYPPIV 160
>emb|CAD29823.2| putative ubiquitin-conjugating enzyme [Populus euramericana]
Length = 160
Score = 316 bits (810), Expect = 3e-85
Identities = 144/157 (91%), Positives = 150/157 (94%)
Frame = -3
Query: 755 GIARGRLTEERKSWRKNHPHGFVAKPETLSDGTINLMVWHCTIPGKTGTDWDGGYFPLTL 576
GIARGRL EERKSWRKNHPHGFVAKPET DGT+NLMVWHCTIPGK GTDW+GGYFPLTL
Sbjct: 4 GIARGRLAEERKSWRKNHPHGFVAKPETQPDGTVNLMVWHCTIPGKLGTDWEGGYFPLTL 63
Query: 575 HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ 396
+FSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ
Sbjct: 64 NFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ 123
Query: 395 PNPADPAQTEGYHLFIQDVAEYKRRVQQQAKLFPPLV 285
PNPADPAQTEGYHLFIQD AEYK+RV+QQAK +P LV
Sbjct: 124 PNPADPAQTEGYHLFIQDAAEYKKRVRQQAKQYPSLV 160
>gb|AAL58113.1|AC092697_1 putative ubiquitin-conjugating enzyme [Oryza sativa (japonica
cultivar-group)]
Length = 160
Score = 308 bits (789), Expect = 7e-83
Identities = 137/157 (87%), Positives = 149/157 (94%)
Frame = -3
Query: 755 GIARGRLTEERKSWRKNHPHGFVAKPETLSDGTINLMVWHCTIPGKTGTDWDGGYFPLTL 576
GIARGRL EERK+WRKNHPHGFVAKPET++DG+ NLM+WHCTIPGK GTDW+GGY+PLTL
Sbjct: 4 GIARGRLAEERKAWRKNHPHGFVAKPETMADGSANLMIWHCTIPGKQGTDWEGGYYPLTL 63
Query: 575 HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ 396
HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ
Sbjct: 64 HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLDQ 123
Query: 395 PNPADPAQTEGYHLFIQDVAEYKRRVQQQAKLFPPLV 285
PNPADPAQT+GYH+FIQD EYKRRV+ QAK +P L+
Sbjct: 124 PNPADPAQTDGYHIFIQDKPEYKRRVRVQAKQYPALL 160
>ref|NP_191346.1| E2 ubiquitin-conjugating-like enzyme, Ahus5; protein id:
At3g57870.1, supported by cDNA: gi_1174161, supported by
cDNA: gi_14596100 [Arabidopsis thaliana]
gi|11272416|pir||T46009 E2 ubiquitin-conjugating-like
enzyme Ahus5 - Arabidopsis thaliana
gi|1174162|gb|AAA86642.1| ubiquitin-conjugating enzyme
gi|3746915|gb|AAC64116.1| E2 ubiquitin-conjugating-like
enzyme [Arabidopsis thaliana] gi|6729530|emb|CAB67615.1|
E2 ubiquitin-conjugating-like enzyme Ahus5 [Arabidopsis
thaliana] gi|14596101|gb|AAK68778.1| E2
ubiquitin-conjugating-like enzyme Ahus5 [Arabidopsis
thaliana] gi|28059255|gb|AAO30040.1| E2
ubiquitin-conjugating-like enzyme Ahus5 [Arabidopsis
thaliana]
Length = 160
Score = 303 bits (776), Expect = 2e-81
Identities = 136/158 (86%), Positives = 146/158 (92%)
Frame = -3
Query: 758 AGIARGRLTEERKSWRKNHPHGFVAKPETLSDGTINLMVWHCTIPGKTGTDWDGGYFPLT 579
+GIARGRL EERKSWRKNHPHGFVAKPET DGT+NLMVWHCTIPGK GTDW+GG+FPLT
Sbjct: 3 SGIARGRLAEERKSWRKNHPHGFVAKPETGQDGTVNLMVWHCTIPGKAGTDWEGGFFPLT 62
Query: 578 LHFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDSGWRPAITVKQILVGIQDLLD 399
+HFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNED GWRPAITVKQILVGIQDLLD
Sbjct: 63 MHFSEDYPSKPPKCKFPQGFFHPNVYPSGTVCLSILNEDYGWRPAITVKQILVGIQDLLD 122
Query: 398 QPNPADPAQTEGYHLFIQDVAEYKRRVQQQAKLFPPLV 285
PNPADPAQT+GYHLF QD EYK+RV+ Q+K +P LV
Sbjct: 123 TPNPADPAQTDGYHLFCQDPVEYKKRVKLQSKQYPALV 160
>pir||T50603 ubiquitin-conjugating enzyme [imported] - Prunus armeniaca
(fragment) gi|2267139|gb|AAB63513.1|
ubiquitin-conjugating enzyme [Prunus armeniaca]
Length = 126
Score = 260 bits (664), Expect = 2e-68
Identities = 116/126 (92%), Positives = 122/126 (96%)
Frame = -3
Query: 662 GTINLMVWHCTIPGKTGTDWDGGYFPLTLHFSEDYPSKPPKCKFPQGFFHPNVYPSGTVC 483
GT+NLMVWHCTIPGKTGTDW+GG+FPLTLHFSEDYPSKPPKCKFP GFFHPNVYPSGTVC
Sbjct: 1 GTVNLMVWHCTIPGKTGTDWEGGFFPLTLHFSEDYPSKPPKCKFPPGFFHPNVYPSGTVC 60
Query: 482 LSILNEDSGWRPAITVKQILVGIQDLLDQPNPADPAQTEGYHLFIQDVAEYKRRVQQQAK 303
LSILNEDSGWRPAITVKQILVGIQDLLDQPNPADPAQTEGYHLFIQD EYK+RV+QQAK
Sbjct: 61 LSILNEDSGWRPAITVKQILVGIQDLLDQPNPADPAQTEGYHLFIQDATEYKKRVRQQAK 120
Query: 302 LFPPLV 285
+PPLV
Sbjct: 121 QYPPLV 126
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 792,193,647
Number of Sequences: 1393205
Number of extensions: 18395239
Number of successful extensions: 47139
Number of sequences better than 10.0: 633
Number of HSP's better than 10.0 without gapping: 44591
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46676
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 46824663816
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)