Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002689A_C01 KMC002689A_c01
(607 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
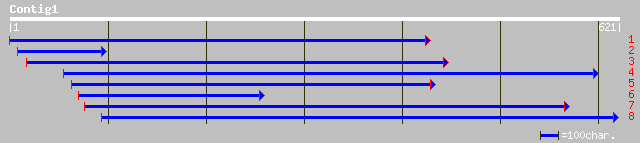
dbj|BAB89792.1| P0677H08.7 [Oryza sativa (japonica cultivar-group)] 128 5e-29
ref|NP_565464.1| expressed protein; protein id: At2g20120.1, sup... 124 7e-28
ref|NP_565465.1| expressed protein; protein id: At2g20130.1, sup... 124 7e-28
gb|AAK95310.1|AF410324_1 At2g20120/T2G17.8 [Arabidopsis thaliana... 121 6e-27
ref|NP_564483.1| expressed protein; protein id: At1g43130.1, sup... 118 5e-26
>dbj|BAB89792.1| P0677H08.7 [Oryza sativa (japonica cultivar-group)]
Length = 259
Score = 128 bits (322), Expect = 5e-29
Identities = 60/71 (84%), Positives = 67/71 (93%)
Frame = -2
Query: 606 PRVGEYAFGFITSTVILQKENEDEELCSVFVPTNHLYIGDILLVNSKDVIRPNLSIREGI 427
PR+GEYAFGFITSTV+LQ + DEELCSV+VPTNHLYIGDI LVNS+++IRPNLSIREGI
Sbjct: 166 PRIGEYAFGFITSTVVLQTDKGDEELCSVYVPTNHLYIGDIFLVNSEEIIRPNLSIREGI 225
Query: 426 EIIVSGGMTMP 394
EIIVSGGMTMP
Sbjct: 226 EIIVSGGMTMP 236
>ref|NP_565464.1| expressed protein; protein id: At2g20120.1, supported by cDNA:
123469., supported by cDNA: gi_15294265, supported by
cDNA: gi_20147284 [Arabidopsis thaliana]
gi|25411966|pir||C84585 hypothetical protein At2g20120
[imported] - Arabidopsis thaliana
gi|4580461|gb|AAD24385.1| expressed protein [Arabidopsis
thaliana] gi|21537202|gb|AAM61543.1| unknown
[Arabidopsis thaliana]
Length = 268
Score = 124 bits (312), Expect = 7e-28
Identities = 59/71 (83%), Positives = 65/71 (91%)
Frame = -2
Query: 606 PRVGEYAFGFITSTVILQKENEDEELCSVFVPTNHLYIGDILLVNSKDVIRPNLSIREGI 427
PRVGEYAFGFITSTV+LQ +EELC V+VPTNHLYIGDILLVNS DVIRPNLS+REGI
Sbjct: 179 PRVGEYAFGFITSTVVLQNYPTEEELCCVYVPTNHLYIGDILLVNSNDVIRPNLSVREGI 238
Query: 426 EIIVSGGMTMP 394
EI+VSGGM+MP
Sbjct: 239 EIVVSGGMSMP 249
>ref|NP_565465.1| expressed protein; protein id: At2g20130.1, supported by cDNA:
12246., supported by cDNA: gi_18253024 [Arabidopsis
thaliana] gi|18253025|gb|AAL62439.1| unknown protein
[Arabidopsis thaliana] gi|20197569|gb|AAD24400.2|
expressed protein [Arabidopsis thaliana]
gi|21537162|gb|AAM61503.1| unknown [Arabidopsis
thaliana] gi|22136466|gb|AAM91311.1| unknown protein
[Arabidopsis thaliana]
Length = 256
Score = 124 bits (312), Expect = 7e-28
Identities = 59/71 (83%), Positives = 65/71 (91%)
Frame = -2
Query: 606 PRVGEYAFGFITSTVILQKENEDEELCSVFVPTNHLYIGDILLVNSKDVIRPNLSIREGI 427
PRVGEYAFGFITSTV+LQ +EELC V+VPTNHLYIGDILLVNS DVIRPNLS+REGI
Sbjct: 169 PRVGEYAFGFITSTVVLQNYPTEEELCCVYVPTNHLYIGDILLVNSNDVIRPNLSVREGI 228
Query: 426 EIIVSGGMTMP 394
EI+VSGGM+MP
Sbjct: 229 EIVVSGGMSMP 239
>gb|AAK95310.1|AF410324_1 At2g20120/T2G17.8 [Arabidopsis thaliana] gi|20147285|gb|AAM10356.1|
At2g20120/T2G17.8 [Arabidopsis thaliana]
Length = 268
Score = 121 bits (304), Expect = 6e-27
Identities = 58/71 (81%), Positives = 64/71 (89%)
Frame = -2
Query: 606 PRVGEYAFGFITSTVILQKENEDEELCSVFVPTNHLYIGDILLVNSKDVIRPNLSIREGI 427
PRVGEYAFGFITSTV+LQ +EELC V+VPTN LYIGDILLVNS DVIRPNLS+REGI
Sbjct: 179 PRVGEYAFGFITSTVVLQNYPTEEELCCVYVPTNRLYIGDILLVNSNDVIRPNLSVREGI 238
Query: 426 EIIVSGGMTMP 394
EI+VSGGM+MP
Sbjct: 239 EIVVSGGMSMP 249
>ref|NP_564483.1| expressed protein; protein id: At1g43130.1, supported by cDNA:
23672. [Arabidopsis thaliana]
gi|12321538|gb|AAG50825.1|AC026757_6 unknown protein
[Arabidopsis thaliana]
Length = 261
Score = 118 bits (296), Expect = 5e-26
Identities = 55/71 (77%), Positives = 66/71 (92%)
Frame = -2
Query: 606 PRVGEYAFGFITSTVILQKENEDEELCSVFVPTNHLYIGDILLVNSKDVIRPNLSIREGI 427
PR+GEYAFGFITS+V LQ ++ +EELCSV+VPTNHLYIGD+ LV+S+++IRPNLSIREGI
Sbjct: 165 PRIGEYAFGFITSSVTLQTDHGEEELCSVYVPTNHLYIGDVFLVSSEEIIRPNLSIREGI 224
Query: 426 EIIVSGGMTMP 394
EIIVS GMTMP
Sbjct: 225 EIIVSVGMTMP 235
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 513,398,133
Number of Sequences: 1393205
Number of extensions: 10913940
Number of successful extensions: 24366
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 23697
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24357
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)