Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002675A_C01 KMC002675A_c01
(629 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
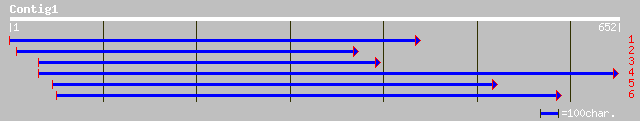
sp|P48488|PP1_MEDVA SERINE/THREONINE PROTEIN PHOSPHATASE PP1 gi|... 218 7e-56
ref|NP_567375.1| protein phosphatase type 1 (AtPP1bg); protein i... 202 4e-51
sp|P48486|PP16_ARATH Serine/threonine protein phosphatase PP1 is... 199 2e-50
pir||T13015 phosphoprotein phosphatase (EC 3.1.3.16) PP1BG - Ara... 195 4e-49
ref|NP_176587.1| phosphoprotein phosphatase-type 1 catalytic sub... 192 2e-48
>sp|P48488|PP1_MEDVA SERINE/THREONINE PROTEIN PHOSPHATASE PP1 gi|1084363|pir||S46282
phosphoprotein phosphatase (EC 3.1.3.16) 1 [similarity]
- alfalfa gi|575672|emb|CAA56766.1| potentially
catalitic subunit of the ser /thr protein phosphatase 1
[Medicago sativa subsp. x varia]
Length = 321
Score = 218 bits (554), Expect = 7e-56
Identities = 102/106 (96%), Positives = 106/106 (99%)
Frame = -1
Query: 629 ENDRGVSFTFGADKVVEFLEHHDLDLICRAHQVVEDGYEFFAKRQLVTVFSAPNYCGEFD 450
ENDRGVSFTFGADKVVEFLEHHDLDLICRAHQVVEDGYEFFAKR+LVTVFSAPNYCGEFD
Sbjct: 215 ENDRGVSFTFGADKVVEFLEHHDLDLICRAHQVVEDGYEFFAKRKLVTVFSAPNYCGEFD 274
Query: 449 NAGAMMSVDDTLTCSFQILKSSEKKGKMGFGNNSSRPGTPPHKGGK 312
NAGAMMSVDD+LTCSFQILKSS+KKGK+GFGNNSSRPGTPPHKGGK
Sbjct: 275 NAGAMMSVDDSLTCSFQILKSSDKKGKVGFGNNSSRPGTPPHKGGK 320
>ref|NP_567375.1| protein phosphatase type 1 (AtPP1bg); protein id: At4g11240.1,
supported by cDNA: 20905., supported by cDNA:
gi_19699315 [Arabidopsis thaliana]
gi|19699316|gb|AAL91268.1| AT4g11240/F8L21_30
[Arabidopsis thaliana] gi|21554190|gb|AAM63269.1|
protein phosphatase type 1 PP1BG [Arabidopsis thaliana]
gi|21689631|gb|AAM67437.1| AT4g11240/F8L21_30
[Arabidopsis thaliana]
Length = 322
Score = 202 bits (513), Expect = 4e-51
Identities = 93/106 (87%), Positives = 100/106 (93%)
Frame = -1
Query: 629 ENDRGVSFTFGADKVVEFLEHHDLDLICRAHQVVEDGYEFFAKRQLVTVFSAPNYCGEFD 450
ENDRGVS+TFGADKV EFL+ HDLDLICRAHQVVEDGYEFFAKRQLVT+FSAPNYCGEFD
Sbjct: 215 ENDRGVSYTFGADKVAEFLQTHDLDLICRAHQVVEDGYEFFAKRQLVTIFSAPNYCGEFD 274
Query: 449 NAGAMMSVDDTLTCSFQILKSSEKKGKMGFGNNSSRPGTPPHKGGK 312
NAGA+MSVDD+LTCSFQILK+SEKKG+ GF NN RPGTPPHKGGK
Sbjct: 275 NAGALMSVDDSLTCSFQILKASEKKGRFGFNNNVPRPGTPPHKGGK 320
>sp|P48486|PP16_ARATH Serine/threonine protein phosphatase PP1 isozyme 6
gi|829255|emb|CAA86339.1| protein phosphatase type 1
[Arabidopsis thaliana] gi|3153203|gb|AAC39460.1|
serine/threonine protein phosphatase type one
[Arabidopsis thaliana]
Length = 322
Score = 199 bits (506), Expect = 2e-50
Identities = 92/106 (86%), Positives = 99/106 (92%)
Frame = -1
Query: 629 ENDRGVSFTFGADKVVEFLEHHDLDLICRAHQVVEDGYEFFAKRQLVTVFSAPNYCGEFD 450
ENDRGVS+TFGADKV EFL+ HDLDLICRAHQV EDGYEFFAKRQLVT+FSAPNYCGEFD
Sbjct: 215 ENDRGVSYTFGADKVAEFLQTHDLDLICRAHQVDEDGYEFFAKRQLVTIFSAPNYCGEFD 274
Query: 449 NAGAMMSVDDTLTCSFQILKSSEKKGKMGFGNNSSRPGTPPHKGGK 312
NAGA+MSVDD+LTCSFQILK+SEKKG+ GF NN RPGTPPHKGGK
Sbjct: 275 NAGALMSVDDSLTCSFQILKASEKKGRFGFNNNVPRPGTPPHKGGK 320
>pir||T13015 phosphoprotein phosphatase (EC 3.1.3.16) PP1BG - Arabidopsis
thaliana gi|5596470|emb|CAB51408.1| protein phosphatase
type 1 PP1BG [Arabidopsis thaliana]
gi|7267823|emb|CAB81225.1| protein phosphatase type 1
PP1BG [Arabidopsis thaliana]
Length = 318
Score = 195 bits (496), Expect = 4e-49
Identities = 90/103 (87%), Positives = 97/103 (93%)
Frame = -1
Query: 629 ENDRGVSFTFGADKVVEFLEHHDLDLICRAHQVVEDGYEFFAKRQLVTVFSAPNYCGEFD 450
ENDRGVS+TFGADKV EFL+ HDLDLICRAHQVVEDGYEFFAKRQLVT+FSAPNYCGEFD
Sbjct: 215 ENDRGVSYTFGADKVAEFLQTHDLDLICRAHQVVEDGYEFFAKRQLVTIFSAPNYCGEFD 274
Query: 449 NAGAMMSVDDTLTCSFQILKSSEKKGKMGFGNNSSRPGTPPHK 321
NAGA+MSVDD+LTCSFQILK+SEKKG+ GF NN RPGTPPHK
Sbjct: 275 NAGALMSVDDSLTCSFQILKASEKKGRFGFNNNVPRPGTPPHK 317
>ref|NP_176587.1| phosphoprotein phosphatase-type 1 catalytic subunit; protein id:
At1g64040.1, supported by cDNA: gi_166798, supported by
cDNA: gi_19698982 [Arabidopsis thaliana]
gi|1346756|sp|P48483|PP13_ARATH Serine/threonine protein
phosphatase PP1 isozyme 3 gi|421852|pir||S31087
phosphoprotein phosphatase (EC 3.1.3.16) 1 (clone TOPP3)
[similarity] - Arabidopsis thaliana
gi|166799|gb|AAA32838.1| phosphoprotein phosphatase 1
gi|19698983|gb|AAL91227.1| putative serine/threonine
protein phosphatase PP1 isozyme 3 [Arabidopsis thaliana]
gi|22136304|gb|AAM91230.1| putative serine/threonine
protein phosphatase PP1 isozyme 3 [Arabidopsis thaliana]
Length = 322
Score = 192 bits (489), Expect = 2e-48
Identities = 90/105 (85%), Positives = 96/105 (90%)
Frame = -1
Query: 629 ENDRGVSFTFGADKVVEFLEHHDLDLICRAHQVVEDGYEFFAKRQLVTVFSAPNYCGEFD 450
ENDRGVS+TFGADKV EFL+ HDLDLICRAHQVVEDGYEFFA RQLVT+FSAPNYCGEFD
Sbjct: 215 ENDRGVSYTFGADKVEEFLQTHDLDLICRAHQVVEDGYEFFANRQLVTIFSAPNYCGEFD 274
Query: 449 NAGAMMSVDDTLTCSFQILKSSEKKGKMGFGNNSSRPGTPPHKGG 315
NAGAMMSVDD+LTCSFQILK+SEKKG GFG N+ R GTPP KGG
Sbjct: 275 NAGAMMSVDDSLTCSFQILKASEKKGNFGFGKNAGRRGTPPRKGG 319
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 562,789,527
Number of Sequences: 1393205
Number of extensions: 12775722
Number of successful extensions: 36691
Number of sequences better than 10.0: 488
Number of HSP's better than 10.0 without gapping: 33694
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36007
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25870486187
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)