Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002665A_C01 KMC002665A_c01
(616 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
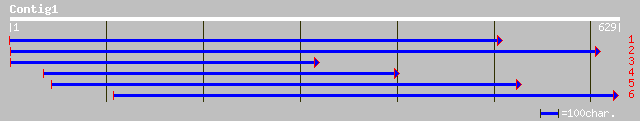
ref|NP_192594.2| hypothetical protein; protein id: At4g08540.1, ... 75 1e-23
gb|AAL59980.1| unknown protein [Arabidopsis thaliana] 75 1e-23
pir||C85085 hypothetical protein AT4g08540 [imported] - Arabidop... 75 1e-23
pir||T01830 hypothetical protein T15F16.11 - Arabidopsis thalian... 75 7e-19
dbj|BAC07067.1| hypothetical protein~similar to Arabidopsis thal... 52 7e-09
>ref|NP_192594.2| hypothetical protein; protein id: At4g08540.1, supported by cDNA:
gi_18176079 [Arabidopsis thaliana]
gi|23297675|gb|AAN13006.1| unknown protein [Arabidopsis
thaliana]
Length = 473
Score = 75.5 bits (184), Expect(2) = 1e-23
Identities = 46/80 (57%), Positives = 56/80 (69%)
Frame = -2
Query: 615 RSVFSLKMARSRTCKQVQQLNKSVWNMDSTISSTTLLESAHSVPPMRIENYMPSSGTSFL 436
RSVFSLKMA SR+ KQ QQLNKS+WN S ISS +LLESAH +P N P+S S+L
Sbjct: 368 RSVFSLKMASSRSGKQAQQLNKSIWNAHSVISS-SLLESAH-LPRNTSYNQDPNSPASYL 425
Query: 435 YATDLSDGKSECLFEGWDIV 376
AT+LS K+ + GWD+V
Sbjct: 426 SATELSTRKNNDM-NGWDLV 444
Score = 56.2 bits (134), Expect(2) = 1e-23
Identities = 23/25 (92%), Positives = 25/25 (100%)
Frame = -1
Query: 361 FPPPPSQSEDVEHWTRAMFIDAKRK 287
+PPPPSQSEDVEHWTRAMFIDAK+K
Sbjct: 449 YPPPPSQSEDVEHWTRAMFIDAKKK 473
>gb|AAL59980.1| unknown protein [Arabidopsis thaliana]
Length = 473
Score = 75.5 bits (184), Expect(2) = 1e-23
Identities = 46/80 (57%), Positives = 56/80 (69%)
Frame = -2
Query: 615 RSVFSLKMARSRTCKQVQQLNKSVWNMDSTISSTTLLESAHSVPPMRIENYMPSSGTSFL 436
RSVFSLKMA SR+ KQ QQLNKS+WN S ISS +LLESAH +P N P+S S+L
Sbjct: 368 RSVFSLKMASSRSGKQAQQLNKSIWNAHSVISS-SLLESAH-LPRNTSYNQDPNSPASYL 425
Query: 435 YATDLSDGKSECLFEGWDIV 376
AT+LS K+ + GWD+V
Sbjct: 426 SATELSTRKNNDM-NGWDLV 444
Score = 56.2 bits (134), Expect(2) = 1e-23
Identities = 23/25 (92%), Positives = 25/25 (100%)
Frame = -1
Query: 361 FPPPPSQSEDVEHWTRAMFIDAKRK 287
+PPPPSQSEDVEHWTRAMFIDAK+K
Sbjct: 449 YPPPPSQSEDVEHWTRAMFIDAKKK 473
>pir||C85085 hypothetical protein AT4g08540 [imported] - Arabidopsis thaliana
gi|7267496|emb|CAB77979.1| hypothetical protein
[Arabidopsis thaliana]
Length = 447
Score = 75.5 bits (184), Expect(2) = 1e-23
Identities = 46/80 (57%), Positives = 56/80 (69%)
Frame = -2
Query: 615 RSVFSLKMARSRTCKQVQQLNKSVWNMDSTISSTTLLESAHSVPPMRIENYMPSSGTSFL 436
RSVFSLKMA SR+ KQ QQLNKS+WN S ISS +LLESAH +P N P+S S+L
Sbjct: 342 RSVFSLKMASSRSGKQAQQLNKSIWNAHSVISS-SLLESAH-LPRNTSYNQDPNSPASYL 399
Query: 435 YATDLSDGKSECLFEGWDIV 376
AT+LS K+ + GWD+V
Sbjct: 400 SATELSTRKNNDM-NGWDLV 418
Score = 56.2 bits (134), Expect(2) = 1e-23
Identities = 23/25 (92%), Positives = 25/25 (100%)
Frame = -1
Query: 361 FPPPPSQSEDVEHWTRAMFIDAKRK 287
+PPPPSQSEDVEHWTRAMFIDAK+K
Sbjct: 423 YPPPPSQSEDVEHWTRAMFIDAKKK 447
>pir||T01830 hypothetical protein T15F16.11 - Arabidopsis thaliana
gi|3377819|gb|AAC28192.1| T15F16.11 gene product
[Arabidopsis thaliana]
Length = 325
Score = 75.1 bits (183), Expect(2) = 7e-19
Identities = 45/80 (56%), Positives = 54/80 (67%)
Frame = -2
Query: 615 RSVFSLKMARSRTCKQVQQLNKSVWNMDSTISSTTLLESAHSVPPMRIENYMPSSGTSFL 436
RSVFSLKMA SR+ KQ QQLNKS+WN S ISS +LLESAH N P+S S+L
Sbjct: 84 RSVFSLKMASSRSGKQAQQLNKSIWNAHSVISS-SLLESAHLPQRNTSYNQDPNSPASYL 142
Query: 435 YATDLSDGKSECLFEGWDIV 376
AT+LS K+ + GWD+V
Sbjct: 143 SATELSTRKNNDM-NGWDLV 161
Score = 40.4 bits (93), Expect(2) = 7e-19
Identities = 15/20 (75%), Positives = 17/20 (85%)
Frame = -1
Query: 361 FPPPPSQSEDVEHWTRAMFI 302
+PPPPSQSEDVEHWT M +
Sbjct: 166 YPPPPSQSEDVEHWTHLMTV 185
>dbj|BAC07067.1| hypothetical protein~similar to Arabidopsis thaliana
chromosome4,At4g08540 [Oryza sativa (japonica
cultivar-group)]
Length = 486
Score = 52.4 bits (124), Expect(2) = 7e-09
Identities = 30/74 (40%), Positives = 48/74 (64%), Gaps = 4/74 (5%)
Frame = -2
Query: 585 SRTCKQVQQLNKSVWNMDSTISS-TTLLESAH-SVPPMRIENYMPSSGTSFLYA--TDLS 418
SR+ KQ QQLN+S+W S ISS ++L++S + ++ P ++N + +S TSFLY+
Sbjct: 381 SRSEKQGQQLNRSIWKASSAISSNSSLMDSVNTAIMPSSLDNLLLNSNTSFLYSGKPTKH 440
Query: 417 DGKSECLFEGWDIV 376
G + + EGWD+V
Sbjct: 441 GGVPDNILEGWDMV 454
Score = 29.3 bits (64), Expect(2) = 7e-09
Identities = 11/16 (68%), Positives = 12/16 (74%)
Frame = -1
Query: 358 PPPPSQSEDVEHWTRA 311
PPPPS+ EDV W RA
Sbjct: 460 PPPPSRVEDVAQWERA 475
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 532,393,899
Number of Sequences: 1393205
Number of extensions: 11792488
Number of successful extensions: 27517
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 26667
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27503
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24854530794
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)