Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002659A_C01 KMC002659A_c01
(750 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
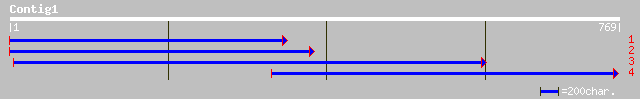
ref|NP_568596.1| CMP-sialic acid transporter-like protein; prote... 209 4e-59
dbj|BAB10651.1| CMP-sialic acid transporter-like protein [Arabid... 209 4e-59
ref|NP_195260.1| UDP-galactose transporter - like protein; prote... 90 3e-17
emb|CAA19645.1| /prediction=(method:''genefinder'', version:''08... 73 5e-12
ref|NP_570019.1| CG2675-PA [Drosophila melanogaster] gi|7290349|... 73 5e-12
>ref|NP_568596.1| CMP-sialic acid transporter-like protein; protein id: At5g41760.1,
supported by cDNA: 103679. [Arabidopsis thaliana]
gi|21536568|gb|AAM60900.1| CMP-sialic acid
transporter-like protein [Arabidopsis thaliana]
gi|28393785|gb|AAO42302.1| putative CMP-sialic acid
transporter [Arabidopsis thaliana]
gi|28973255|gb|AAO63952.1| putative CMP-sialic acid
transporter [Arabidopsis thaliana]
Length = 340
Score = 209 bits (532), Expect(2) = 4e-59
Identities = 100/135 (74%), Positives = 116/135 (85%)
Frame = -3
Query: 688 FGTIFNLARLVVDDFRGGFENGPWWQRIFNGYTITTWLVVLNLGSTGLLVSWLMKYADNI 509
FG++FN+ARL+ DDFR GFE GPWWQRIF+GY+ITTWLVVLNLGSTGLLVSWLMKYADNI
Sbjct: 206 FGSLFNVARLIADDFRHGFEKGPWWQRIFDGYSITTWLVVLNLGSTGLLVSWLMKYADNI 265
Query: 508 VKVYSTSMAMLLTMVLSIFLFDFKPTLQLFLGIVICMMSLHMYFAPSKMLLDMPLAVKPD 329
VKVYSTSMAMLLTMV SI+LF FKPTLQLFLGIVIC+MSLHMYFAP L+D+P+ +
Sbjct: 266 VKVYSTSMAMLLTMVASIYLFSFKPTLQLFLGIVICIMSLHMYFAPPHTLVDLPVTNEAH 325
Query: 328 EEKLIEDSVDRRTHS 284
+ L + V+ +T S
Sbjct: 326 AKTLKQVVVEEKTDS 340
Score = 41.6 bits (96), Expect(2) = 4e-59
Identities = 16/19 (84%), Positives = 19/19 (99%)
Frame = -2
Query: 749 FLMKKNSDTLYWQNIQLYT 693
FLMK+N+DTLYWQN+QLYT
Sbjct: 187 FLMKRNNDTLYWQNLQLYT 205
>dbj|BAB10651.1| CMP-sialic acid transporter-like protein [Arabidopsis thaliana]
Length = 323
Score = 209 bits (532), Expect(2) = 4e-59
Identities = 100/135 (74%), Positives = 116/135 (85%)
Frame = -3
Query: 688 FGTIFNLARLVVDDFRGGFENGPWWQRIFNGYTITTWLVVLNLGSTGLLVSWLMKYADNI 509
FG++FN+ARL+ DDFR GFE GPWWQRIF+GY+ITTWLVVLNLGSTGLLVSWLMKYADNI
Sbjct: 189 FGSLFNVARLIADDFRHGFEKGPWWQRIFDGYSITTWLVVLNLGSTGLLVSWLMKYADNI 248
Query: 508 VKVYSTSMAMLLTMVLSIFLFDFKPTLQLFLGIVICMMSLHMYFAPSKMLLDMPLAVKPD 329
VKVYSTSMAMLLTMV SI+LF FKPTLQLFLGIVIC+MSLHMYFAP L+D+P+ +
Sbjct: 249 VKVYSTSMAMLLTMVASIYLFSFKPTLQLFLGIVICIMSLHMYFAPPHTLVDLPVTNEAH 308
Query: 328 EEKLIEDSVDRRTHS 284
+ L + V+ +T S
Sbjct: 309 AKTLKQVVVEEKTDS 323
Score = 41.6 bits (96), Expect(2) = 4e-59
Identities = 16/19 (84%), Positives = 19/19 (99%)
Frame = -2
Query: 749 FLMKKNSDTLYWQNIQLYT 693
FLMK+N+DTLYWQN+QLYT
Sbjct: 170 FLMKRNNDTLYWQNLQLYT 188
>ref|NP_195260.1| UDP-galactose transporter - like protein; protein id: At4g35340.1
[Arabidopsis thaliana] gi|7488414|pir||T06123
UDP-galactose transport protein homolog F23E12.100 -
Arabidopsis thaliana gi|3080416|emb|CAA18735.1|
UDP-galactose transporter-like protein [Arabidopsis
thaliana] gi|7270486|emb|CAB80251.1| UDP-galactose
transporter-like protein [Arabidopsis thaliana]
Length = 102
Score = 90.1 bits (222), Expect = 3e-17
Identities = 47/101 (46%), Positives = 67/101 (65%)
Frame = -3
Query: 676 FNLARLVVDDFRGGFENGPWWQRIFNGYTITTWLVVLNLGSTGLLVSWLMKYADNIVKVY 497
FN +V+ DF G F+GY+ T L++LN +G+ VS +MKYADNIVKVY
Sbjct: 3 FNAVAIVIQDFDAVANKG-----FFHGYSFITLLMILNHALSGIAVSMVMKYADNIVKVY 57
Query: 496 STSMAMLLTMVLSIFLFDFKPTLQLFLGIVICMMSLHMYFA 374
STS+AMLLT V+S+FLF+F +L FLG + +S++++ A
Sbjct: 58 STSVAMLLTAVVSVFLFNFHLSLAFFLGSTVVSVSVYLHSA 98
>emb|CAA19645.1| /prediction=(method:''genefinder'', version:''084'',
score:''73.05'')~/match=(desc:''UDP-GALACTOSE
TRANSLOCATOR (UDP-GALACTOSE TRANSPORTER) (UGT) (UDP-GAL-
TR)'', species:''Homo sapiens (Human)'',
ranges:(query:9322..9444,
target:SWISS-PROT::P78381:75..35, score:''108.00''),
(query:8468..8827, target:SWISS-PROT::P78381:189..70,
score:''311.00''), (query:7973..8401,
target:SWISS-PROT::P78381:338..196, score:''395.00'')),
method:''blastx'',
version:''1.4.9'')~/match=(desc:''CMP-SIALIC ACID
TRANSPORTER (CMP->
Length = 368
Score = 72.8 bits (177), Expect = 5e-12
Identities = 44/123 (35%), Positives = 67/123 (53%), Gaps = 2/123 (1%)
Frame = -3
Query: 676 FNLARLVVDDFRGGFENGPWWQRIFNGYTITTWLVVLNLGSTGLLVSWLMKYADNIVKVY 497
F L V+D F+ G F GY + W +VL GL+V+ ++KYADNI+K +
Sbjct: 248 FGLLTCFVNDGSRIFDQG-----FFKGYDLFVWYLVLLQAGGGLIVAVVVKYADNILKGF 302
Query: 496 STSMAMLLTMVLSIFLFDFKPTLQLFLGIVICMMSLHMYFAPSKMLLDMPLAVKP--DEE 323
+TS+A++++ V SI++FDF TLQ G + + S+ +Y P P DEE
Sbjct: 303 ATSLAIIISCVASIYIFDFNLTLQFSFGAGLVIASIFLYGYDPARSAPKPTMHGPGGDEE 362
Query: 322 KLI 314
KL+
Sbjct: 363 KLL 365
>ref|NP_570019.1| CG2675-PA [Drosophila melanogaster] gi|7290349|gb|AAF45808.1|
CG2675-PA [Drosophila melanogaster]
gi|15128553|dbj|BAB62747.1| UDP-galactose transporter
[Drosophila melanogaster]
gi|15213693|gb|AAK92124.1|AF397530_1 CMP-sialic
acid/UDP-galactose transporter [Drosophila melanogaster]
gi|27819743|gb|AAO24924.1| SD16302p [Drosophila
melanogaster]
Length = 357
Score = 72.8 bits (177), Expect = 5e-12
Identities = 44/123 (35%), Positives = 67/123 (53%), Gaps = 2/123 (1%)
Frame = -3
Query: 676 FNLARLVVDDFRGGFENGPWWQRIFNGYTITTWLVVLNLGSTGLLVSWLMKYADNIVKVY 497
F L V+D F+ G F GY + W +VL GL+V+ ++KYADNI+K +
Sbjct: 237 FGLLTCFVNDGSRIFDQG-----FFKGYDLFVWYLVLLQAGGGLIVAVVVKYADNILKGF 291
Query: 496 STSMAMLLTMVLSIFLFDFKPTLQLFLGIVICMMSLHMYFAPSKMLLDMPLAVKP--DEE 323
+TS+A++++ V SI++FDF TLQ G + + S+ +Y P P DEE
Sbjct: 292 ATSLAIIISCVASIYIFDFNLTLQFSFGAGLVIASIFLYGYDPARSAPKPTMHGPGGDEE 351
Query: 322 KLI 314
KL+
Sbjct: 352 KLL 354
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 643,720,375
Number of Sequences: 1393205
Number of extensions: 13915064
Number of successful extensions: 34075
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 32725
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34041
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36314099463
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)