Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002648A_C01 KMC002648A_c01
(505 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
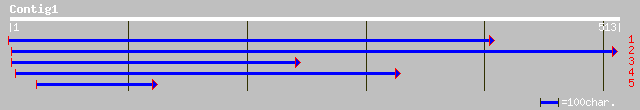
ref|NP_179126.1| putative lysosomal acid lipase; protein id: At2... 67 5e-27
dbj|BAB08297.1| contains similarity to lipase~gene_id:MUA22.18 [... 39 2e-07
ref|NP_568295.1| putative protein; protein id: At5g14180.1, supp... 39 2e-07
gb|EAA05393.1| ebiP3881 [Anopheles gambiae str. PEST] 43 1e-04
gb|AAM34340.1| similar to Mus musculus (Mouse). Adult male tongu... 40 1e-04
>ref|NP_179126.1| putative lysosomal acid lipase; protein id: At2g15230.1
[Arabidopsis thaliana] gi|25411617|pir||E84526 probable
lysosomal acid lipase [imported] - Arabidopsis thaliana
gi|4585908|gb|AAD25569.1| putative lysosomal acid lipase
[Arabidopsis thaliana]
Length = 344
Score = 67.4 bits (163), Expect(3) = 5e-27
Identities = 30/47 (63%), Positives = 34/47 (71%)
Frame = -2
Query: 504 FYLEQEPHPSSSTHLCHLLQMIRKGTFCMYDYGKLKNLIEYGSIKPP 364
+YL+ EPHPSS ++ HL QMIRKGTF YDYG KNL YG KPP
Sbjct: 218 YYLDYEPHPSSVKNIRHLFQMIRKGTFAQYDYGYFKNLRTYGLSKPP 264
Score = 61.2 bits (147), Expect(3) = 5e-27
Identities = 26/45 (57%), Positives = 33/45 (72%)
Frame = -3
Query: 377 VSNHPHFDLSRIPKSLPLWMAYGGNDALADISDFQHTLKDLQSTP 243
+S P F LS IP SLP+WM YGG D LAD++D +HTL +L S+P
Sbjct: 260 LSKPPEFILSHIPASLPMWMGYGGTDGLADVTDVEHTLAELPSSP 304
Score = 33.9 bits (76), Expect(3) = 5e-27
Identities = 12/20 (60%), Positives = 17/20 (85%)
Frame = -1
Query: 244 PEVVYLENYGHVDFILSLQA 185
PE++YLE+YGH+DF+L A
Sbjct: 304 PELLYLEDYGHIDFVLGSSA 323
>dbj|BAB08297.1| contains similarity to lipase~gene_id:MUA22.18 [Arabidopsis
thaliana]
Length = 318
Score = 39.3 bits (90), Expect(3) = 2e-07
Identities = 16/34 (47%), Positives = 24/34 (70%)
Frame = -3
Query: 365 PHFDLSRIPKSLPLWMAYGGNDALADISDFQHTL 264
P +++S IP LPL+ +YGG D+LAD+ D + L
Sbjct: 239 PAYNISAIPHELPLFFSYGGLDSLADVKDVEFLL 272
Score = 33.9 bits (76), Expect(3) = 2e-07
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Frame = -2
Query: 501 YLEQEPHPSSSTHLCHLLQMIRKGTFCMYDYGKL-KNLIEYGSIKPP 364
+L EP +S+ ++ HL Q +R Y+YG +N+ YG PP
Sbjct: 193 FLANEPQSTSTKNMIHLAQTVRDKELRKYNYGSSDRNIKHYGQAIPP 239
Score = 21.9 bits (45), Expect(3) = 2e-07
Identities = 7/18 (38%), Positives = 13/18 (71%)
Frame = -1
Query: 238 VVYLENYGHVDFILSLQA 185
V ++++Y H DFI+ + A
Sbjct: 285 VQFVKDYAHADFIMGVTA 302
>ref|NP_568295.1| putative protein; protein id: At5g14180.1, supported by cDNA:
gi_15081687, supported by cDNA: gi_20147186 [Arabidopsis
thaliana] gi|15081688|gb|AAK82499.1| AT5g14180/MUA22_18
[Arabidopsis thaliana] gi|20147187|gb|AAM10310.1|
AT5g14180/MUA22_18 [Arabidopsis thaliana]
Length = 205
Score = 39.3 bits (90), Expect(3) = 2e-07
Identities = 16/34 (47%), Positives = 24/34 (70%)
Frame = -3
Query: 365 PHFDLSRIPKSLPLWMAYGGNDALADISDFQHTL 264
P +++S IP LPL+ +YGG D+LAD+ D + L
Sbjct: 126 PAYNISAIPHELPLFFSYGGLDSLADVKDVEFLL 159
Score = 33.9 bits (76), Expect(3) = 2e-07
Identities = 16/47 (34%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Frame = -2
Query: 501 YLEQEPHPSSSTHLCHLLQMIRKGTFCMYDYGKL-KNLIEYGSIKPP 364
+L EP +S+ ++ HL Q +R Y+YG +N+ YG PP
Sbjct: 80 FLANEPQSTSTKNMIHLAQTVRDKELRKYNYGSSDRNIKHYGQAIPP 126
Score = 21.9 bits (45), Expect(3) = 2e-07
Identities = 7/18 (38%), Positives = 13/18 (71%)
Frame = -1
Query: 238 VVYLENYGHVDFILSLQA 185
V ++++Y H DFI+ + A
Sbjct: 172 VQFVKDYAHADFIMGVTA 189
>gb|EAA05393.1| ebiP3881 [Anopheles gambiae str. PEST]
Length = 774
Score = 42.7 bits (99), Expect = 0.002
Identities = 20/45 (44%), Positives = 24/45 (52%)
Frame = -2
Query: 498 LEQEPHPSSSTHLCHLLQMIRKGTFCMYDYGKLKNLIEYGSIKPP 364
LE P +S L H Q G F YD+G NLI YGS++PP
Sbjct: 242 LENAPAGASVNQLVHYAQGYNSGRFRQYDFGLTLNLIRYGSVRPP 286
Score = 40.8 bits (94), Expect(2) = 1e-04
Identities = 20/45 (44%), Positives = 23/45 (50%)
Frame = -2
Query: 498 LEQEPHPSSSTHLCHLLQMIRKGTFCMYDYGKLKNLIEYGSIKPP 364
L P +S L H Q G F +DYG NLI YGSI+PP
Sbjct: 656 LANTPAGASVNQLVHYAQGYNSGRFRQFDYGLTLNLIRYGSIRPP 700
Score = 25.4 bits (54), Expect(2) = 1e-04
Identities = 13/42 (30%), Positives = 23/42 (53%)
Frame = -3
Query: 365 PHFDLSRIPKSLPLWMAYGGNDALADISDFQHTLKDLQSTPG 240
P + L R+ + P+ + YG ND LA +SD + +++ G
Sbjct: 700 PDYPLDRV--TAPVALHYGDNDWLAAVSDVRQLHSSIRNPIG 739
>gb|AAM34340.1| similar to Mus musculus (Mouse). Adult male tongue cDNA, RIKEN
full-length enriched library, clone:2310079O20, full
insert sequence [Dictyostelium discoideum]
Length = 429
Score = 40.4 bits (93), Expect(2) = 1e-04
Identities = 18/49 (36%), Positives = 26/49 (52%)
Frame = -2
Query: 504 FYLEQEPHPSSSTHLCHLLQMIRKGTFCMYDYGKLKNLIEYGSIKPPTL 358
F EP +S ++ H Q++ F YDYG + NL+ YG KPP +
Sbjct: 301 FVSGNEPGGTSLRNMVHFTQLVNSKQFQHYDYGVIGNLLHYGHEKPPLI 349
Score = 25.8 bits (55), Expect(2) = 1e-04
Identities = 10/25 (40%), Positives = 16/25 (64%), Gaps = 4/25 (16%)
Frame = -1
Query: 247 PPEVVY----LENYGHVDFILSLQA 185
PPE + +ENY H+DF+ ++ A
Sbjct: 384 PPETILSWDIIENYAHLDFVWAIDA 408
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 461,712,034
Number of Sequences: 1393205
Number of extensions: 9576452
Number of successful extensions: 20971
Number of sequences better than 10.0: 36
Number of HSP's better than 10.0 without gapping: 20510
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20968
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15362785481
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)