Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002642A_C01 KMC002642A_c01
(624 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
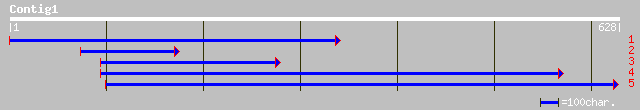
dbj|BAB86894.1| syringolide-induced protein B13-1-9 [Glycine max] 175 5e-43
gb|AAK44147.2|AF370332_1 putative harpin-induced protein [Arabid... 95 8e-19
ref|NP_196250.1| harpin-induced protein-like; protein id: At5g06... 95 8e-19
gb|AAF88023.1|AF264699_1 NDR1/HIN1-like protein 3 [Arabidopsis t... 88 8e-17
gb|AAF62403.1|AF212183_1 harpin inducing protein [Nicotiana taba... 87 1e-16
>dbj|BAB86894.1| syringolide-induced protein B13-1-9 [Glycine max]
Length = 203
Score = 175 bits (443), Expect = 5e-43
Identities = 90/125 (72%), Positives = 105/125 (84%)
Frame = -2
Query: 623 KLNIYYDKVEALAFYEDSRFASDDKVITHMNSFRQYKKSSSPMSATFSGQHVMVLDSDQL 444
KL+IYYDKVEALAFYED+RFA+ D VITHMNSFRQYKKS+SPMSA FSG+ V++L+S+Q+
Sbjct: 81 KLSIYYDKVEALAFYEDARFANYD-VITHMNSFRQYKKSTSPMSAVFSGKKVLMLNSEQV 139
Query: 443 SELSKDKSAHVYDIYVKLHFRIRFRLGDFIFNNDLKPKVKCDLKVPLDSNATFTFFAVTK 264
S+L++DKS VYDIYVKL+FRIRFRLGD I N LKPKVKC LKVP + TFT F TK
Sbjct: 140 SKLNQDKSDGVYDIYVKLNFRIRFRLGDSISGN-LKPKVKCHLKVPFSKSGTFTLFETTK 198
Query: 263 CDVDF 249
C V F
Sbjct: 199 CSVHF 203
>gb|AAK44147.2|AF370332_1 putative harpin-induced protein [Arabidopsis thaliana]
Length = 223
Score = 94.7 bits (234), Expect = 8e-19
Identities = 49/126 (38%), Positives = 77/126 (60%), Gaps = 1/126 (0%)
Frame = -2
Query: 623 KLNIYYDKVEALAFYEDSRFASDDKVITHMNSFRQYKKSSSPMSATFSGQHVMVLDSDQL 444
++ +YYD++E +Y D RF + + + F Q K+++ + GQ +++LD +
Sbjct: 103 RIGVYYDEIEVRGYYGDQRFGMSNNI----SKFYQGHKNTTVVGTKLVGQQLVLLDGGER 158
Query: 443 SELSKDKSAHVYDIYVKLHFRIRFRLGDFIFNNDLKPKVKCDLKVPLDSNATFTF-FAVT 267
+L++D ++ +Y I KL +IRF+ G I + KPK+KCDLKVPL SN+T F F T
Sbjct: 159 KDLNEDVNSQIYRIDAKLRLKIRFKFG-LIKSWRFKPKIKCDLKVPLTSNSTSGFVFQPT 217
Query: 266 KCDVDF 249
KCDVDF
Sbjct: 218 KCDVDF 223
>ref|NP_196250.1| harpin-induced protein-like; protein id: At5g06320.1, supported by
cDNA: gi_9502175 [Arabidopsis thaliana]
gi|9758412|dbj|BAB08954.1| harpin-induced protein-like
[Arabidopsis thaliana] gi|24030178|gb|AAN41271.1|
putative harpin-induced protein [Arabidopsis thaliana]
Length = 231
Score = 94.7 bits (234), Expect = 8e-19
Identities = 49/126 (38%), Positives = 77/126 (60%), Gaps = 1/126 (0%)
Frame = -2
Query: 623 KLNIYYDKVEALAFYEDSRFASDDKVITHMNSFRQYKKSSSPMSATFSGQHVMVLDSDQL 444
++ +YYD++E +Y D RF + + + F Q K+++ + GQ +++LD +
Sbjct: 111 RIGVYYDEIEVRGYYGDQRFGMSNNI----SKFYQGHKNTTVVGTKLVGQQLVLLDGGER 166
Query: 443 SELSKDKSAHVYDIYVKLHFRIRFRLGDFIFNNDLKPKVKCDLKVPLDSNATFTF-FAVT 267
+L++D ++ +Y I KL +IRF+ G I + KPK+KCDLKVPL SN+T F F T
Sbjct: 167 KDLNEDVNSQIYRIDAKLRLKIRFKFG-LIKSWRFKPKIKCDLKVPLTSNSTSGFVFQPT 225
Query: 266 KCDVDF 249
KCDVDF
Sbjct: 226 KCDVDF 231
>gb|AAF88023.1|AF264699_1 NDR1/HIN1-like protein 3 [Arabidopsis thaliana]
gi|24417302|gb|AAN60261.1| unknown [Arabidopsis
thaliana]
Length = 230
Score = 88.2 bits (217), Expect = 8e-17
Identities = 48/126 (38%), Positives = 76/126 (60%), Gaps = 1/126 (0%)
Frame = -2
Query: 623 KLNIYYDKVEALAFYEDSRFASDDKVITHMNSFRQYKKSSSPMSATFSGQHVMVLDSDQL 444
++ +YY+ +E +Y D RF + + + F Q K+++ + GQ +++LD +
Sbjct: 111 RIGVYYE-IEVRGYYGDQRFGMSNNI----SKFYQGHKNTTVVGTKLVGQQLVLLDGGER 165
Query: 443 SELSKDKSAHVYDIYVKLHFRIRFRLGDFIFNNDLKPKVKCDLKVPLDSNATFTF-FAVT 267
+L++D ++ +Y I KL +IRF+ G I + KPK+KCDLKVPL SN+T F F T
Sbjct: 166 KDLNEDVNSQIYRIDAKLRLKIRFKFG-LIKSWRFKPKIKCDLKVPLTSNSTSGFVFQPT 224
Query: 266 KCDVDF 249
KCDVDF
Sbjct: 225 KCDVDF 230
>gb|AAF62403.1|AF212183_1 harpin inducing protein [Nicotiana tabacum]
gi|22830759|dbj|BAC15623.1| hin1 [Nicotiana tabacum]
Length = 229
Score = 87.4 bits (215), Expect = 1e-16
Identities = 47/127 (37%), Positives = 78/127 (61%), Gaps = 2/127 (1%)
Frame = -2
Query: 623 KLNIYYDKVEALAFYEDSRFASDDKVITHMNSFRQYKKSSSPMSATFSGQHVMVLDSDQL 444
++ IYYD +EA A Y+ RF S T++ F Q K++S + F GQ +++L +
Sbjct: 109 RIGIYYDSIEARALYQGERFDS-----TNLEPFYQGHKNTSSLRPVFKGQSLVLLGDREK 163
Query: 443 SELSKDKSAHVYDIYVKLHFRIRFRLGDFIFNNDLKPKVKCDLKVPLDSN--ATFTFFAV 270
S + +K+ VY++ VKL+ RIR ++G +I + +KPK++CD KVPL SN ++ F
Sbjct: 164 SNYNNEKNLGVYEMEVKLYMRIRLKVG-WIKTHKIKPKIECDFKVPLGSNGRSSSANFEE 222
Query: 269 TKCDVDF 249
T+C +D+
Sbjct: 223 TRCHLDW 229
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 472,886,114
Number of Sequences: 1393205
Number of extensions: 9798147
Number of successful extensions: 38084
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 32383
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37582
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)