Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002623A_C01 KMC002623A_c01
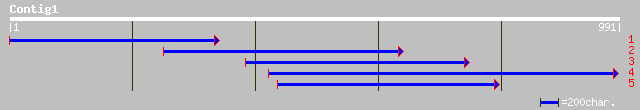
(913 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM76754.1| hypothetical protein [Arabidopsis thaliana] 69 1e-10
dbj|BAC43251.1| unknown protein [Arabidopsis thaliana] gi|284167... 69 1e-10
ref|NP_181004.1| hypothetical protein; protein id: At2g34570.1 [... 56 7e-07
ref|NP_115710.1| hypothetical protein MGC14595 [Homo sapiens] gi... 42 0.017
gb|AAC98238.1| NADH dehydrogenase subunit 4 [Haemonchus placei] ... 37 0.32
>gb|AAM76754.1| hypothetical protein [Arabidopsis thaliana]
Length = 281
Score = 68.9 bits (167), Expect = 1e-10
Identities = 38/74 (51%), Positives = 48/74 (64%), Gaps = 10/74 (13%)
Frame = -1
Query: 787 RSITSRNKTDIKDKPQFKRKRAKGPNPLSCKKKKSHENP--------NIGSMKEMK--GD 638
R ++++N +KD+PQFKR RAKGPNPLSC KKK ENP N + KE K G
Sbjct: 202 RVVSTKNGLGVKDRPQFKRNRAKGPNPLSCMKKKK-ENPQSKSKADSNSNAQKEKKEGGS 260
Query: 637 DTVRRSRKRTRSRK 596
DT +RSRKR++ K
Sbjct: 261 DTQKRSRKRSKKGK 274
>dbj|BAC43251.1| unknown protein [Arabidopsis thaliana] gi|28416751|gb|AAO42906.1|
At2g34570 [Arabidopsis thaliana]
Length = 82
Score = 68.9 bits (167), Expect = 1e-10
Identities = 38/74 (51%), Positives = 48/74 (64%), Gaps = 10/74 (13%)
Frame = -1
Query: 787 RSITSRNKTDIKDKPQFKRKRAKGPNPLSCKKKKSHENP--------NIGSMKEMK--GD 638
R ++++N +KD+PQFKR RAKGPNPLSC KKK ENP N + KE K G
Sbjct: 3 RVVSTKNGLGVKDRPQFKRNRAKGPNPLSCMKKKK-ENPQSKSKADSNSNAQKEKKEGGS 61
Query: 637 DTVRRSRKRTRSRK 596
DT +RSRKR++ K
Sbjct: 62 DTQKRSRKRSKKGK 75
>ref|NP_181004.1| hypothetical protein; protein id: At2g34570.1 [Arabidopsis
thaliana] gi|25408336|pir||C84758 hypothetical protein
At2g34570 [imported] - Arabidopsis thaliana
gi|3128208|gb|AAC26688.1| hypothetical protein
[Arabidopsis thaliana]
Length = 258
Score = 56.2 bits (134), Expect = 7e-07
Identities = 27/55 (49%), Positives = 36/55 (65%)
Frame = -1
Query: 787 RSITSRNKTDIKDKPQFKRKRAKGPNPLSCKKKKSHENPNIGSMKEMKGDDTVRR 623
R ++++N +KD+PQFKR RAKGPNPLSC KKK ENP S + + +R
Sbjct: 202 RVVSTKNGLGVKDRPQFKRNRAKGPNPLSCMKKKK-ENPQSKSKADSNSNAQKQR 255
>ref|NP_115710.1| hypothetical protein MGC14595 [Homo sapiens]
gi|13543606|gb|AAH05955.1|AAH05955 Similar to RIKEN cDNA
D530033C11 gene [Homo sapiens]
gi|18490658|gb|AAH22441.1| hypothetical protein MGC14595
[Homo sapiens]
Length = 249
Score = 41.6 bits (96), Expect = 0.017
Identities = 23/49 (46%), Positives = 25/49 (50%)
Frame = -1
Query: 736 KRKRAKGPNPLSCKKKKSHENPNIGSMKEMKGDDTVRRSRKRTRSRKRP 590
KRK+ GPNPLSC KKK S E RR RKR R+R P
Sbjct: 195 KRKKISGPNPLSCLKKKKKAPDTQSSASEK------RRKRKRIRNRSNP 237
>gb|AAC98238.1| NADH dehydrogenase subunit 4 [Haemonchus placei]
gi|4063264|gb|AAC98239.1| NADH dehydrogenase subunit 4
[Haemonchus placei] gi|4063272|gb|AAC98243.1| NADH
dehydrogenase subunit 4 [Haemonchus placei]
Length = 153
Score = 37.4 bits (85), Expect = 0.32
Identities = 32/129 (24%), Positives = 57/129 (43%), Gaps = 7/129 (5%)
Frame = -1
Query: 508 LLLLVNEERSKFSMMAWRGKMHISCTFEFYIIEVYQRNINGHIVFFCISYYLPNCYFFIV 329
+L++++ + S F MM G + T FY+I + + + +++F S+ + F IV
Sbjct: 7 ILMMMSSKNSAFMMMLAHG---YTSTLMFYVIGEFYHSSSSRMIYFMNSFMNSSMIFSIV 63
Query: 328 EVF*Y-------PNLSNPRTLLCLFN*CPCRLAKSVLNFSFCCFFGSIFVMSFLFVISLA 170
F + P+LS + + N L+K + F F F S + FL V S A
Sbjct: 64 FAFIFLSNSGMPPSLSFLSEFIIIVN--SMMLSKILFFFIFLYFVISFYYSLFLIVNSFA 121
Query: 169 YNHFFARGN 143
++ N
Sbjct: 122 GKYYLKFNN 130
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 754,707,157
Number of Sequences: 1393205
Number of extensions: 16578717
Number of successful extensions: 40313
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 37577
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40004
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 49918505760
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)