Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002615A_C01 KMC002615A_c01
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
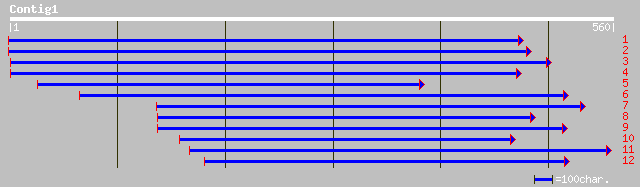
ref|NP_172889.1| protein kinase; protein id: At1g14370.1, suppor... 67 2e-10
ref|NP_178383.1| putative protein kinase; protein id: At2g02800.... 64 1e-09
dbj|BAC42058.1| putative protein kinase [Arabidopsis thaliana] 64 1e-09
gb|AAK92662.1|AC090487_4 Putative serine /threonine kinase simil... 62 6e-09
gb|AAL84315.1|AC073556_32 putative protein tyrosine-serine-threo... 47 2e-04
>ref|NP_172889.1| protein kinase; protein id: At1g14370.1, supported by cDNA:
gi_12248034, supported by cDNA: gi_16649048, supported
by cDNA: gi_2852446 [Arabidopsis thaliana]
gi|25287626|pir||T52285 serine/threonine-specific
protein kinase APK2a (EC 2.7.1.-) [imported] -
Arabidopsis thaliana gi|2852447|dbj|BAA24694.1| protein
kinase [Arabidopsis thaliana]
gi|7262679|gb|AAF43937.1|AC012188_14 Strong similarity,
practically identical, to APK2a protein from Arabidopsis
thaliana gb|D88206 and contains a Eukaryotic protein
kinase PF|00069 domain. ESTs gb|AA712684, gb|H76755,
gb|AA651227 come from this gene
gi|12248035|gb|AAG50109.1|AF334731_1 putative protein
kinase [Arabidopsis thaliana] gi|16649049|gb|AAL24376.1|
Strong similarity to APK2a protein [Arabidopsis
thaliana]
Length = 426
Score = 66.6 bits (161), Expect = 2e-10
Identities = 45/93 (48%), Positives = 58/93 (61%), Gaps = 6/93 (6%)
Frame = -1
Query: 557 AFMAATLALQCLNNEAKARPPMTQVLETLEQIEA-----PKNAGRNSLAQHHNNRFQ-TP 396
AF AA LALQCLN +AK RP M++VL TLEQ+E+ K+ S HH++ Q +P
Sbjct: 338 AFTAANLALQCLNPDAKLRPKMSEVLVTLEQLESVAKPGTKHTQMESPRFHHSSVMQKSP 397
Query: 395 VRRSPARNRTPLNLTPTASPLPSPLPAHRHSPR 297
VR S +R L++TP ASPLPS + SPR
Sbjct: 398 VRYS--HDRPLLHMTPGASPLPS----YTQSPR 424
>ref|NP_178383.1| putative protein kinase; protein id: At2g02800.1, supported by
cDNA: gi_16226562, supported by cDNA: gi_2852448
[Arabidopsis thaliana] gi|7434289|pir||T00848 probable
serine/threonine-specific protein kinase T20F6.6 (EC
2.7.1.-) - Arabidopsis thaliana
gi|2852449|dbj|BAA24695.1| protein kinase [Arabidopsis
thaliana] gi|2947061|gb|AAC05342.1| putative protein
kinase [Arabidopsis thaliana]
gi|16226563|gb|AAL16201.1|AF428432_1 At2g02800/T20F6.6
[Arabidopsis thaliana] gi|21928081|gb|AAM78069.1|
At2g02800/T20F6.6 [Arabidopsis thaliana]
Length = 426
Score = 64.3 bits (155), Expect = 1e-09
Identities = 43/94 (45%), Positives = 59/94 (62%), Gaps = 7/94 (7%)
Frame = -1
Query: 557 AFMAATLALQCLNNEAKARPPMTQVLETLEQIEA--PKNAGRNSLAQHHNNRFQ--TPVR 390
A+ AA+LALQCLN +AK RP M++VL L+Q+E+ P N AQ + R + V+
Sbjct: 335 AYTAASLALQCLNPDAKLRPKMSEVLAKLDQLESTKPGTGVGNRQAQIDSPRGSNGSIVQ 394
Query: 389 RSPAR---NRTPLNLTPTASPLPSPLPAHRHSPR 297
+SP R +R L++TP ASPLP+ H HSPR
Sbjct: 395 KSPRRYSYDRPLLHITPGASPLPT----HNHSPR 424
>dbj|BAC42058.1| putative protein kinase [Arabidopsis thaliana]
Length = 426
Score = 64.3 bits (155), Expect = 1e-09
Identities = 43/94 (45%), Positives = 59/94 (62%), Gaps = 7/94 (7%)
Frame = -1
Query: 557 AFMAATLALQCLNNEAKARPPMTQVLETLEQIEA--PKNAGRNSLAQHHNNRFQ--TPVR 390
A+ AA+LALQCLN +AK RP M++VL L+Q+E+ P N AQ + R + V+
Sbjct: 335 AYTAASLALQCLNPDAKLRPKMSEVLAKLDQLESTKPGTGVGNRQAQIDSPRGSNGSIVQ 394
Query: 389 RSPAR---NRTPLNLTPTASPLPSPLPAHRHSPR 297
+SP R +R L++TP ASPLP+ H HSPR
Sbjct: 395 KSPRRYSYDRPLLHITPGASPLPT----HNHSPR 424
>gb|AAK92662.1|AC090487_4 Putative serine /threonine kinase similar to NAK [Oryza sativa
(japonica cultivar-group)]
gi|15451543|gb|AAK98667.1|AC021893_1 Putative
serine/threonine-specific kinase [Oryza sativa]
Length = 428
Score = 61.6 bits (148), Expect = 6e-09
Identities = 38/87 (43%), Positives = 51/87 (57%), Gaps = 4/87 (4%)
Frame = -1
Query: 557 AFMAATLALQCLNNEAKARPPMTQVLETLEQIEAPKNAGRNSLAQHHNNRFQTPVRRSPA 378
A AT+ALQC+ +EAK RP M++VLE L+Q++ PK + V RSP
Sbjct: 337 AHAIATIALQCIRSEAKMRPQMSEVLEKLQQLQDPKYNVTSPQVDTRRRSSSGSVPRSPM 396
Query: 377 RNR-TPLNLTPTASPLP---SPLPAHR 309
R + +P L+ +ASPLP SPLPA R
Sbjct: 397 RMQPSPRRLSASASPLPAAGSPLPACR 423
>gb|AAL84315.1|AC073556_32 putative protein tyrosine-serine-threonine kinase [Oryza sativa
(japonica cultivar-group)]
Length = 422
Score = 47.0 bits (110), Expect = 2e-04
Identities = 32/87 (36%), Positives = 48/87 (54%), Gaps = 3/87 (3%)
Frame = -1
Query: 557 AFMAATLALQCLNNEAKARPPMTQVLETLEQIEAPK---NAGRNSLAQHHNNRFQTPVRR 387
A A +ALQC+ N+AK RP M++VLE LEQ++ K + + + + N ++P+R
Sbjct: 339 AHAIANIALQCICNDAKMRPRMSEVLEELEQLQDSKYNMASPQVDIRRTSNAVPKSPMRI 398
Query: 386 SPARNRTPLNLTPTASPLPSPLPAHRH 306
P+ R +L ASPLP A H
Sbjct: 399 QPSPRR---SLGAAASPLPGYRTAKVH 422
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 483,015,410
Number of Sequences: 1393205
Number of extensions: 10512449
Number of successful extensions: 39943
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 34449
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 38959
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)