Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002612A_C01 KMC002612A_c01
(667 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
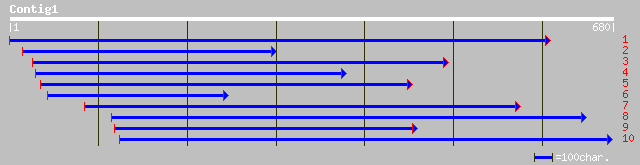
dbj|BAB39968.1| putative peptidyl-prolyl cis-trans isomerase, ch... 209 3e-53
sp|Q41651|CYPB_VICFA Peptidyl-prolyl cis-trans isomerase, chloro... 204 1e-51
ref|NP_191762.1| peptidylprolyl isomerase ROC4; protein id: At3g... 201 7e-51
gb|AAM63944.1| peptidylprolyl isomerase ROC4 [Arabidopsis thaliana] 201 7e-51
gb|AAG40378.1|AF325026_1 AT3g62030 [Arabidopsis thaliana] 199 2e-50
>dbj|BAB39968.1| putative peptidyl-prolyl cis-trans isomerase, chloroplast precursor
[Oryza sativa (japonica cultivar-group)]
gi|13486749|dbj|BAB39983.1| putative peptidyl-prolyl
cis-trans isomerase, chloroplast precursor [Oryza sativa
(japonica cultivar-group)] gi|15408836|dbj|BAB64228.1|
putative peptidyl-prolyl cis-trans isomerase,
chloroplast precursor [Oryza sativa (japonica
cultivar-group)]
Length = 231
Score = 209 bits (532), Expect = 3e-53
Identities = 95/117 (81%), Positives = 106/117 (90%)
Frame = -2
Query: 666 SFHRIIQEFMIQGGDFTEGDGTGGTSIYGSSFEDESFALKHVGPGVLSMANAGPNTNGSQ 487
SFHRII++FMIQGGDF +GTGG SIYG F+DE+F LKH GPGVLSMANAGP+TNGSQ
Sbjct: 114 SFHRIIKDFMIQGGDFQNNNGTGGRSIYGECFDDENFTLKHTGPGVLSMANAGPDTNGSQ 173
Query: 486 FFICTVKTPWLDNRHVVFGHVIDGMDVVRTLESQETSRLDIPRKPCRIVNCGELPQD 316
FFICTVKTPWLDNRHVVFGHV++GMDVV+ LESQETSR DIP++PCRIVNCGELP D
Sbjct: 174 FFICTVKTPWLDNRHVVFGHVLEGMDVVKNLESQETSRSDIPKQPCRIVNCGELPVD 230
>sp|Q41651|CYPB_VICFA Peptidyl-prolyl cis-trans isomerase, chloroplast precursor (PPIase)
(Rotamase) (Cyclophilin) (Cyclosporin A-binding protein)
(CYP B) gi|7437324|pir||T12096 peptidylprolyl isomerase
(EC 5.2.1.8) - fava bean gi|499693|gb|AAA64430.1|
cyclophilin
Length = 248
Score = 204 bits (518), Expect = 1e-51
Identities = 97/117 (82%), Positives = 104/117 (87%), Gaps = 1/117 (0%)
Frame = -2
Query: 663 FHRIIQEFMIQGGDFTEGDGTGGTSIYGSSFEDESFALKHVGPGVLSMANAGPNTNGSQF 484
FHRII FMIQGGDFTEG+GTGG SIYGS FEDESF LKHVGPGVLSMANAGPNTNGSQF
Sbjct: 131 FHRIIPNFMIQGGDFTEGNGTGGVSIYGSKFEDESFDLKHVGPGVLSMANAGPNTNGSQF 190
Query: 483 FICTVKTPWLDNRHVVFGHVIDGMDVVRTLESQETSRLD-IPRKPCRIVNCGELPQD 316
FICTV TPWLDNRHVVFGHVI+G+DVV+ LESQETS+LD P+KPC+I GELP D
Sbjct: 191 FICTVPTPWLDNRHVVFGHVIEGLDVVKQLESQETSKLDNSPKKPCKIAKSGELPLD 247
>ref|NP_191762.1| peptidylprolyl isomerase ROC4; protein id: At3g62030.1, supported
by cDNA: 29220., supported by cDNA: gi_13358215,
supported by cDNA: gi_16648946, supported by cDNA:
gi_20259871, supported by cDNA: gi_405130 [Arabidopsis
thaliana] gi|461899|sp|P34791|CYP4_ARATH Peptidyl-prolyl
cis-trans isomerase, chloroplast precursor (PPIase)
(Rotamase) (Cyclophilin) (Cyclosporin A-binding protein)
gi|1076368|pir||B53422 peptidylprolyl isomerase (EC
5.2.1.8) ROC4 - Arabidopsis thaliana
gi|405131|gb|AAA20048.1| cyclophilin
gi|1322278|gb|AAB96831.1| cyclophilin [Arabidopsis
thaliana] gi|6899901|emb|CAB71910.1| peptidylprolyl
isomerase ROC4 [Arabidopsis thaliana]
gi|16648947|gb|AAL24325.1| peptidylprolyl isomerase ROC4
[Arabidopsis thaliana] gi|20259872|gb|AAM13283.1|
peptidylprolyl isomerase ROC4 [Arabidopsis thaliana]
Length = 260
Score = 201 bits (511), Expect = 7e-51
Identities = 92/117 (78%), Positives = 103/117 (87%)
Frame = -2
Query: 666 SFHRIIQEFMIQGGDFTEGDGTGGTSIYGSSFEDESFALKHVGPGVLSMANAGPNTNGSQ 487
SFHRII++FMIQGGDFTEG+GTGG SIYG+ FEDE+F LKH GPG+LSMANAGPNTNGSQ
Sbjct: 143 SFHRIIKDFMIQGGDFTEGNGTGGISIYGAKFEDENFTLKHTGPGILSMANAGPNTNGSQ 202
Query: 486 FFICTVKTPWLDNRHVVFGHVIDGMDVVRTLESQETSRLDIPRKPCRIVNCGELPQD 316
FFICTVKT WLDN+HVVFG VI+GM +VRTLESQET D+P+K CRI CGELP D
Sbjct: 203 FFICTVKTSWLDNKHVVFGQVIEGMKLVRTLESQETRAFDVPKKGCRIYACGELPLD 259
>gb|AAM63944.1| peptidylprolyl isomerase ROC4 [Arabidopsis thaliana]
Length = 260
Score = 201 bits (511), Expect = 7e-51
Identities = 92/117 (78%), Positives = 103/117 (87%)
Frame = -2
Query: 666 SFHRIIQEFMIQGGDFTEGDGTGGTSIYGSSFEDESFALKHVGPGVLSMANAGPNTNGSQ 487
SFHRII++FMIQGGDFTEG+GTGG SIYG+ FEDE+F LKH GPG+LSMANAGPNTNGSQ
Sbjct: 143 SFHRIIKDFMIQGGDFTEGNGTGGISIYGAKFEDENFTLKHTGPGILSMANAGPNTNGSQ 202
Query: 486 FFICTVKTPWLDNRHVVFGHVIDGMDVVRTLESQETSRLDIPRKPCRIVNCGELPQD 316
FFICTVKT WLDN+HVVFG VI+GM +VRTLESQET D+P+K CRI CGELP D
Sbjct: 203 FFICTVKTSWLDNKHVVFGQVIEGMKLVRTLESQETRAFDVPKKGCRIYACGELPLD 259
>gb|AAG40378.1|AF325026_1 AT3g62030 [Arabidopsis thaliana]
Length = 260
Score = 199 bits (507), Expect = 2e-50
Identities = 91/117 (77%), Positives = 103/117 (87%)
Frame = -2
Query: 666 SFHRIIQEFMIQGGDFTEGDGTGGTSIYGSSFEDESFALKHVGPGVLSMANAGPNTNGSQ 487
SFHRII++FMIQGGDFTEG+GTGG SIYG+ FEDE+F LKH GPG+LSMANAGPNTNGS+
Sbjct: 143 SFHRIIKDFMIQGGDFTEGNGTGGISIYGAKFEDENFTLKHTGPGILSMANAGPNTNGSK 202
Query: 486 FFICTVKTPWLDNRHVVFGHVIDGMDVVRTLESQETSRLDIPRKPCRIVNCGELPQD 316
FFICTVKT WLDN+HVVFG VI+GM +VRTLESQET D+P+K CRI CGELP D
Sbjct: 203 FFICTVKTSWLDNKHVVFGQVIEGMKLVRTLESQETRAFDVPKKGCRIYACGELPLD 259
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 552,940,836
Number of Sequences: 1393205
Number of extensions: 11599503
Number of successful extensions: 27205
Number of sequences better than 10.0: 818
Number of HSP's better than 10.0 without gapping: 24654
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26621
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)