Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002605A_C02 KMC002605A_c02
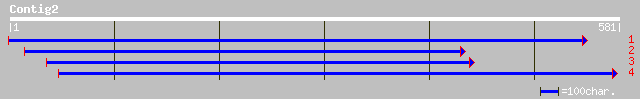
(581 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_300494.1| ClpC protease [Chlamydophila pneumoniae J138] g... 34 1.5
ref|NP_444865.1| ATP-dependent Clp protease, ATP-binding subunit... 34 1.5
ref|XP_146743.1| similar to olfactory receptor MOR165-3 [Mus mus... 33 3.4
ref|NP_224637.1| ClpC Protease [Chlamydophila pneumoniae CWL029]... 33 3.4
ref|XP_236095.1| similar to olfactory receptor MOR165-7 [Mus mus... 32 4.4
>ref|NP_300494.1| ClpC protease [Chlamydophila pneumoniae J138]
gi|25289886|pir||C86545 ClpC proteinase [imported] -
Chlamydophila pneumoniae (strain J138)
gi|8978809|dbj|BAA98645.1| ClpC protease [Chlamydophila
pneumoniae J138]
Length = 845
Score = 33.9 bits (76), Expect = 1.5
Identities = 27/73 (36%), Positives = 33/73 (44%), Gaps = 1/73 (1%)
Frame = +2
Query: 362 IRHQNDSV*STKRKNADQHLPNFRK-ILKPRHQIAYRAPQNCSISR*SSDKNPQKSSPRL 538
I HQ+DSV +N RK ILK + P + S S SS NP S L
Sbjct: 113 ILHQSDSVALQVLENLHIDPREVRKEILKELETFNLQLPPSSSSSSSSSRSNPSSSKSPL 172
Query: 539 WHSVMIDKRSKLN 577
HS+ DK KL+
Sbjct: 173 GHSLGSDKNEKLS 185
>ref|NP_444865.1| ATP-dependent Clp protease, ATP-binding subunit [Chlamydophila
pneumoniae AR39] gi|14194545|sp|Q9Z8A6|CLPC_CHLPN
Probable ATP-dependent CLP protease ATP-binding subunit
gi|11265233|pir||G81590 ATP-dependent Clp proteinase,
ATP-binding chain CP0316 [imported] - Chlamydophila
pneumoniae (strain AR39) gi|7189244|gb|AAF38172.1|
ATP-dependent Clp protease, ATP-binding subunit
[Chlamydophila pneumoniae AR39]
Length = 845
Score = 33.9 bits (76), Expect = 1.5
Identities = 27/73 (36%), Positives = 33/73 (44%), Gaps = 1/73 (1%)
Frame = +2
Query: 362 IRHQNDSV*STKRKNADQHLPNFRK-ILKPRHQIAYRAPQNCSISR*SSDKNPQKSSPRL 538
I HQ+DSV +N RK ILK + P + S S SS NP S L
Sbjct: 113 ILHQSDSVALQVLENLHIDPREVRKEILKELETFNLQLPPSSSSSSSSSRSNPSSSKSPL 172
Query: 539 WHSVMIDKRSKLN 577
HS+ DK KL+
Sbjct: 173 GHSLGSDKNEKLS 185
>ref|XP_146743.1| similar to olfactory receptor MOR165-3 [Mus musculus]
gi|25032241|ref|XP_205679.1| similar to olfactory
receptor MOR165-3 [Mus musculus]
Length = 312
Score = 32.7 bits (73), Expect = 3.4
Identities = 27/111 (24%), Positives = 47/111 (42%), Gaps = 12/111 (10%)
Frame = -3
Query: 408 AFLRLVLYTESF*WRMMLFLRDCSILLWGLLYFIVGGRSVHTCCIICITL--SGTIAFII 235
A+ R V + + L + C L+ G + G +HT CI+ +T TI
Sbjct: 119 AYDRYVAICNPLLYNVTLSSKVCCYLMLGSYFMGFSGAMIHTGCILRLTFCDGNTINHYF 178
Query: 234 CN-------TCDRNILGGVFLPLLNGLCLL---LVIFSRYFLALLAALKIK 112
C+ +C + + L ++ G ++ ++IF Y L + LKIK
Sbjct: 179 CDLLPLLQISCTSTYINEIELFIVAGKDIIVPTIIIFISYGFILFSVLKIK 229
>ref|NP_224637.1| ClpC Protease [Chlamydophila pneumoniae CWL029]
gi|7428218|pir||G72079 endopeptidase Clp (EC 3.4.21.-)
ATP-binding chain clpC [similarity] - Chlamydophila
pneumoniae (strain CWL029) gi|4376719|gb|AAD18581.1|
ClpC Protease [Chlamydophila pneumoniae CWL029]
Length = 845
Score = 32.7 bits (73), Expect = 3.4
Identities = 26/73 (35%), Positives = 33/73 (44%), Gaps = 1/73 (1%)
Frame = +2
Query: 362 IRHQNDSV*STKRKNADQHLPNFRK-ILKPRHQIAYRAPQNCSISR*SSDKNPQKSSPRL 538
I HQ+DSV +N RK IL+ + P + S S SS NP S L
Sbjct: 113 ILHQSDSVALQVLENLHIDPREVRKEILRELETFNLQLPPSSSSSSSSSRSNPSSSKSPL 172
Query: 539 WHSVMIDKRSKLN 577
HS+ DK KL+
Sbjct: 173 GHSLGSDKNEKLS 185
>ref|XP_236095.1| similar to olfactory receptor MOR165-7 [Mus musculus] [Rattus
norvegicus]
Length = 310
Score = 32.3 bits (72), Expect = 4.4
Identities = 28/111 (25%), Positives = 47/111 (42%), Gaps = 12/111 (10%)
Frame = -3
Query: 408 AFLRLVLYTESF*WRMMLFLRDCSILLWGLLYFIVGGRSVHTCCIICITL--SGTIAFII 235
A+ R V + + L + C L+ G + G +HT CI+ +T TI
Sbjct: 119 AYDRYVAICNPLLYNVTLSSKVCFYLMLGSYFMGFSGAMIHTGCILRLTFCDGNTINHYF 178
Query: 234 CN-------TCDRNILGGVFLPLLNGLCLL---LVIFSRYFLALLAALKIK 112
C+ +C + + L ++ G ++ L+IF Y L + LKIK
Sbjct: 179 CDLLPLLQISCTSTYVNEIELFIVAGKDIIVPTLIIFISYGFILFSILKIK 229
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 493,128,932
Number of Sequences: 1393205
Number of extensions: 10879027
Number of successful extensions: 30492
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 29637
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30475
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)