Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002598A_C01 KMC002598A_c01
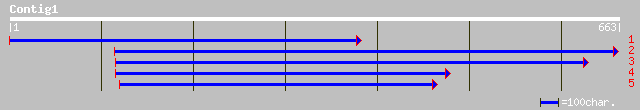
(653 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T49961 hypothetical protein F8M21.150 - Arabidopsis thalian... 117 1e-25
ref|NP_197030.2| putative protein; protein id: At5g15260.1, supp... 117 1e-25
ref|NP_566133.1| Expressed protein; protein id: At3g01170.1, sup... 103 3e-21
gb|AAF26157.1|AC008261_14 unknown protein [Arabidopsis thaliana] 82 8e-15
ref|NP_187269.1| unknown protein; protein id: At3g06180.1 [Arabi... 64 1e-09
>pir||T49961 hypothetical protein F8M21.150 - Arabidopsis thaliana
gi|7671495|emb|CAB89336.1| putative protein [Arabidopsis
thaliana]
Length = 294
Score = 117 bits (294), Expect = 1e-25
Identities = 61/74 (82%), Positives = 64/74 (86%), Gaps = 1/74 (1%)
Frame = -2
Query: 652 IQLETEDCVKNSPSLSKDGGDV-KKGLFELPRDHHRELEAELKKMAPPNGRAVLILRARC 476
IQLETEDCVK+S S G V KKGLFELPRDHHRELEAELKKMAPPNGRAVL+ RA+C
Sbjct: 139 IQLETEDCVKSSNS----GKSVSKKGLFELPRDHHRELEAELKKMAPPNGRAVLVFRAKC 194
Query: 475 GCSVGRLEVPGHKK 434
GCSVGRLEVPG KK
Sbjct: 195 GCSVGRLEVPGPKK 208
>ref|NP_197030.2| putative protein; protein id: At5g15260.1, supported by cDNA:
gi_18086572 [Arabidopsis thaliana]
gi|18086573|gb|AAL57710.1| AT5g15260/F8M21_150
[Arabidopsis thaliana] gi|21689601|gb|AAM67422.1|
AT5g15260/F8M21_150 [Arabidopsis thaliana]
Length = 234
Score = 117 bits (294), Expect = 1e-25
Identities = 61/74 (82%), Positives = 64/74 (86%), Gaps = 1/74 (1%)
Frame = -2
Query: 652 IQLETEDCVKNSPSLSKDGGDV-KKGLFELPRDHHRELEAELKKMAPPNGRAVLILRARC 476
IQLETEDCVK+S S G V KKGLFELPRDHHRELEAELKKMAPPNGRAVL+ RA+C
Sbjct: 155 IQLETEDCVKSSNS----GKSVSKKGLFELPRDHHRELEAELKKMAPPNGRAVLVFRAKC 210
Query: 475 GCSVGRLEVPGHKK 434
GCSVGRLEVPG KK
Sbjct: 211 GCSVGRLEVPGPKK 224
>ref|NP_566133.1| Expressed protein; protein id: At3g01170.1, supported by cDNA:
105104. [Arabidopsis thaliana]
Length = 215
Score = 103 bits (256), Expect = 3e-21
Identities = 53/79 (67%), Positives = 61/79 (77%)
Frame = -2
Query: 652 IQLETEDCVKNSPSLSKDGGDVKKGLFELPRDHHRELEAELKKMAPPNGRAVLILRARCG 473
IQLETE+CVK+S + K+G+FELPR HHRELEAELKKMAPPNGRAVL+ RARCG
Sbjct: 144 IQLETEECVKSSSN--------KRGMFELPRVHHRELEAELKKMAPPNGRAVLVFRARCG 195
Query: 472 CSVGRLEVPGHKKPVGRIE 416
CSV RL V G KK +I+
Sbjct: 196 CSVRRLVVSGPKKQQRKIK 214
>gb|AAF26157.1|AC008261_14 unknown protein [Arabidopsis thaliana]
Length = 596
Score = 81.6 bits (200), Expect = 8e-15
Identities = 41/58 (70%), Positives = 47/58 (80%)
Frame = -2
Query: 652 IQLETEDCVKNSPSLSKDGGDVKKGLFELPRDHHRELEAELKKMAPPNGRAVLILRAR 479
IQLETE+CVK+S + K+G+FELPR HHRELEAELKKMAPPNGRAVL+ RAR
Sbjct: 144 IQLETEECVKSSSN--------KRGMFELPRVHHRELEAELKKMAPPNGRAVLVFRAR 193
>ref|NP_187269.1| unknown protein; protein id: At3g06180.1 [Arabidopsis thaliana]
gi|6862922|gb|AAF30311.1|AC018907_11 unknown protein
[Arabidopsis thaliana]
Length = 241
Score = 64.3 bits (155), Expect = 1e-09
Identities = 35/73 (47%), Positives = 47/73 (63%)
Frame = -2
Query: 652 IQLETEDCVKNSPSLSKDGGDVKKGLFELPRDHHRELEAELKKMAPPNGRAVLILRARCG 473
+QL+TE+CVK+ + D K G P + L AEL++MAPPNGRAVL+ R+RCG
Sbjct: 151 LQLQTEECVKSGATKEIDRLPWKGGSESNP--DYECLRAELRRMAPPNGRAVLLFRSRCG 208
Query: 472 CSVGRLEVPGHKK 434
C V +LE G K+
Sbjct: 209 CPVAKLEGWGPKR 221
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 541,603,743
Number of Sequences: 1393205
Number of extensions: 11547871
Number of successful extensions: 24822
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 23793
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24744
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 28144814643
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)