Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002587A_C01 KMC002587A_c01
(621 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
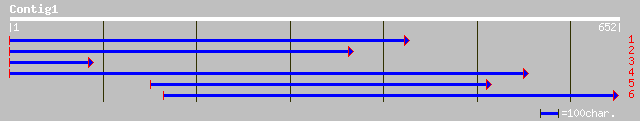
sp|P52780|SYQ_LUPLU GLUTAMINYL-TRNA SYNTHETASE (GLUTAMINE--TRNA ... 197 7e-50
dbj|BAB03361.1| unnamed protein product [Oryza sativa (japonica ... 174 8e-43
pir||D86383 probable glutaminyl-tRNA synthetase (glnrs) [importe... 164 7e-40
gb|AAG28806.2|AC079374_9 tRNA-glutamine synthetase, putative [Ar... 164 7e-40
ref|NP_173906.2| tRNA-glutamine synthetase, putative; protein id... 164 7e-40
>sp|P52780|SYQ_LUPLU GLUTAMINYL-TRNA SYNTHETASE (GLUTAMINE--TRNA LIGASE) (GLNRS)
gi|7437754|pir||T09643 glutamine-tRNA ligase (EC
6.1.1.18) - yellow lupine gi|2995455|emb|CAA62901.1|
tRNA-glutamine synthetase [Lupinus luteus]
Length = 794
Score = 197 bits (502), Expect = 7e-50
Identities = 95/114 (83%), Positives = 103/114 (90%)
Frame = -3
Query: 619 YDPSKKTKPKGVLHWVAQPSSGVEPLKVEVRLFERLFLSENPAELENWLGDLNPHSKVVI 440
YDPSKKTKPKGVLHWV+QPS GV+PLKVEVRLFERLFLSENPAEL+NWLGDLNPHSKV I
Sbjct: 681 YDPSKKTKPKGVLHWVSQPSPGVDPLKVEVRLFERLFLSENPAELDNWLGDLNPHSKVEI 740
Query: 439 PNAYGGSSLQGSKVGDSFPFERLGYFVVDKDSSSEKLVFNRTVTLKDSYTKGGK 278
NAYG S L+ +K+GD F FERLGYF VD+DS+ EKLVFNRTVTLKDSY KGGK
Sbjct: 741 SNAYGVSLLKDAKLGDRFQFERLGYFAVDQDSTPEKLVFNRTVTLKDSYGKGGK 794
>dbj|BAB03361.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 791
Score = 174 bits (441), Expect = 8e-43
Identities = 84/113 (74%), Positives = 94/113 (82%)
Frame = -3
Query: 619 YDPSKKTKPKGVLHWVAQPSSGVEPLKVEVRLFERLFLSENPAELENWLGDLNPHSKVVI 440
YDPSK TKPKGVLHWVAQP+ GVEPLKVEVRLF++LFLSENPAELE+WLGDLNP+SK VI
Sbjct: 677 YDPSKTTKPKGVLHWVAQPAPGVEPLKVEVRLFDKLFLSENPAELEDWLGDLNPNSKEVI 736
Query: 439 PNAYGGSSLQGSKVGDSFPFERLGYFVVDKDSSSEKLVFNRTVTLKDSYTKGG 281
AY SL + +GD F FERLGYF VD DS+ E +VFNRTVTL+DSY K G
Sbjct: 737 KGAYAVPSLATAVLGDKFQFERLGYFAVDTDSTPENIVFNRTVTLRDSYGKAG 789
>pir||D86383 probable glutaminyl-tRNA synthetase (glnrs) [imported] -
Arabidopsis thaliana
Length = 729
Score = 164 bits (416), Expect = 7e-40
Identities = 81/115 (70%), Positives = 94/115 (81%), Gaps = 1/115 (0%)
Frame = -3
Query: 619 YDPSKKTKPKGVLHWVAQPSSGVEPLKVEVRLFERLFLSENPAELEN-WLGDLNPHSKVV 443
YDP KK+KPKGVLHWVA+ S G EP+KVEVRLFE+LF SENPAEL + WL D+NP+SK+V
Sbjct: 615 YDPEKKSKPKGVLHWVAESSPGEEPIKVEVRLFEKLFNSENPAELNDAWLTDINPNSKMV 674
Query: 442 IPNAYGGSSLQGSKVGDSFPFERLGYFVVDKDSSSEKLVFNRTVTLKDSYTKGGK 278
I AY S+L+ + VGD F FERLGY+ VDKDS KLVFNRTVTL+DSY KGGK
Sbjct: 675 ISGAYAVSTLKDAAVGDRFQFERLGYYAVDKDSEPGKLVFNRTVTLRDSYGKGGK 729
>gb|AAG28806.2|AC079374_9 tRNA-glutamine synthetase, putative [Arabidopsis thaliana]
Length = 786
Score = 164 bits (416), Expect = 7e-40
Identities = 81/115 (70%), Positives = 94/115 (81%), Gaps = 1/115 (0%)
Frame = -3
Query: 619 YDPSKKTKPKGVLHWVAQPSSGVEPLKVEVRLFERLFLSENPAELEN-WLGDLNPHSKVV 443
YDP KK+KPKGVLHWVA+ S G EP+KVEVRLFE+LF SENPAEL + WL D+NP+SK+V
Sbjct: 672 YDPEKKSKPKGVLHWVAESSPGEEPIKVEVRLFEKLFNSENPAELNDAWLTDINPNSKMV 731
Query: 442 IPNAYGGSSLQGSKVGDSFPFERLGYFVVDKDSSSEKLVFNRTVTLKDSYTKGGK 278
I AY S+L+ + VGD F FERLGY+ VDKDS KLVFNRTVTL+DSY KGGK
Sbjct: 732 ISGAYAVSTLKDAAVGDRFQFERLGYYAVDKDSEPGKLVFNRTVTLRDSYGKGGK 786
>ref|NP_173906.2| tRNA-glutamine synthetase, putative; protein id: At1g25350.1,
supported by cDNA: gi_17065035 [Arabidopsis thaliana]
gi|17065036|gb|AAL32672.1| Unknown protein [Arabidopsis
thaliana] gi|21387137|gb|AAM47972.1| unknown protein
[Arabidopsis thaliana]
Length = 795
Score = 164 bits (416), Expect = 7e-40
Identities = 81/115 (70%), Positives = 94/115 (81%), Gaps = 1/115 (0%)
Frame = -3
Query: 619 YDPSKKTKPKGVLHWVAQPSSGVEPLKVEVRLFERLFLSENPAELEN-WLGDLNPHSKVV 443
YDP KK+KPKGVLHWVA+ S G EP+KVEVRLFE+LF SENPAEL + WL D+NP+SK+V
Sbjct: 681 YDPEKKSKPKGVLHWVAESSPGEEPIKVEVRLFEKLFNSENPAELNDAWLTDINPNSKMV 740
Query: 442 IPNAYGGSSLQGSKVGDSFPFERLGYFVVDKDSSSEKLVFNRTVTLKDSYTKGGK 278
I AY S+L+ + VGD F FERLGY+ VDKDS KLVFNRTVTL+DSY KGGK
Sbjct: 741 ISGAYAVSTLKDAAVGDRFQFERLGYYAVDKDSEPGKLVFNRTVTLRDSYGKGGK 795
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 540,206,425
Number of Sequences: 1393205
Number of extensions: 11752747
Number of successful extensions: 25761
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 24756
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25565
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)