Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002585A_C01 KMC002585A_c01
(681 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
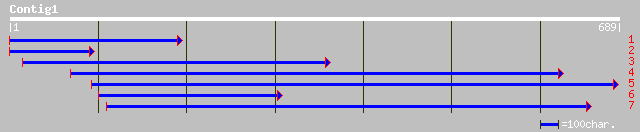
ref|NP_683383.1| unknown protein; protein id: At1g45196.1 [Arabi... 73 4e-12
gb|AAK76561.1| unknown protein [Arabidopsis thaliana] 55 7e-07
ref|NP_568012.1| expressed protein; protein id: At4g36970.1 [Ara... 55 7e-07
ref|NP_027421.1| expressed protein; protein id: At2g02170.1, sup... 51 2e-05
gb|AAO15296.1| Unknown protein [Oryza sativa (japonica cultivar-... 44 0.003
>ref|NP_683383.1| unknown protein; protein id: At1g45196.1 [Arabidopsis thaliana]
Length = 515
Score = 72.8 bits (177), Expect = 4e-12
Identities = 40/72 (55%), Positives = 54/72 (74%), Gaps = 3/72 (4%)
Frame = -2
Query: 680 EMKLEKKRASSMDKIMNKLRLAQTKAQEMRSSALANQAHQVTRTSH-KVISFRRTG--QM 510
+MKLEKKR+SSM+KIM K++ A+ +A+EMR S L N +V+ SH K SF+R+G ++
Sbjct: 447 QMKLEKKRSSSMEKIMRKVKSAEKRAEEMRRSVLDN---RVSTASHGKASSFKRSGKKKI 503
Query: 509 GSLSGCFTCHAF 474
SLSGCFTCH F
Sbjct: 504 PSLSGCFTCHVF 515
>gb|AAK76561.1| unknown protein [Arabidopsis thaliana]
Length = 427
Score = 55.5 bits (132), Expect = 7e-07
Identities = 32/71 (45%), Positives = 47/71 (66%), Gaps = 7/71 (9%)
Frame = -2
Query: 680 EMKLEKKRASSMDKIMNKLRLAQTKAQEMRSSALANQ------AHQVTRTSHKVISF-RR 522
E+KLEKK+++SMDKI+NKL+ A+ KAQEMR S+++++ HQ++R S K+ RR
Sbjct: 344 EVKLEKKKSASMDKILNKLQTAKIKAQEMRRSSVSSEHEQQQGNHQISRNSVKITHLVRR 403
Query: 521 TGQMGSLSGCF 489
M CF
Sbjct: 404 HTFMTPFMTCF 414
>ref|NP_568012.1| expressed protein; protein id: At4g36970.1 [Arabidopsis thaliana]
gi|25407782|pir||F85436 hypothetical protein AT4g36970
[imported] - Arabidopsis thaliana
gi|4006876|emb|CAB16794.1| hypothetical protein
[Arabidopsis thaliana] gi|7270646|emb|CAB80363.1|
hypothetical protein [Arabidopsis thaliana]
gi|23297309|gb|AAN12938.1| unknown protein [Arabidopsis
thaliana]
Length = 427
Score = 55.5 bits (132), Expect = 7e-07
Identities = 32/71 (45%), Positives = 47/71 (66%), Gaps = 7/71 (9%)
Frame = -2
Query: 680 EMKLEKKRASSMDKIMNKLRLAQTKAQEMRSSALANQ------AHQVTRTSHKVISF-RR 522
E+KLEKK+++SMDKI+NKL+ A+ KAQEMR S+++++ HQ++R S K+ RR
Sbjct: 344 EVKLEKKKSASMDKILNKLQTAKIKAQEMRRSSVSSEHEQQQGNHQISRNSVKITHLVRR 403
Query: 521 TGQMGSLSGCF 489
M CF
Sbjct: 404 HTFMTPFMTCF 414
>ref|NP_027421.1| expressed protein; protein id: At2g02170.1, supported by cDNA:
gi_14326557 [Arabidopsis thaliana]
gi|25410968|pir||G84433 hypothetical protein At2g02170
[imported] - Arabidopsis thaliana
gi|4038035|gb|AAC97217.1| expressed protein [Arabidopsis
thaliana] gi|14326558|gb|AAK60323.1|AF385733_1
At2g02170/F5O4.6 [Arabidopsis thaliana]
gi|27764932|gb|AAO23587.1| At2g02170/F5O4.6 [Arabidopsis
thaliana]
Length = 486
Score = 50.8 bits (120), Expect = 2e-05
Identities = 25/69 (36%), Positives = 43/69 (62%)
Frame = -2
Query: 680 EMKLEKKRASSMDKIMNKLRLAQTKAQEMRSSALANQAHQVTRTSHKVISFRRTGQMGSL 501
E+K+E+ + + D++M KL + KA+E R++A A + HQ +T + RRTG++ SL
Sbjct: 418 EVKVERIKGRAQDRLMKKLATIERKAEEKRAAAEAKKDHQAAKTEKQAEQIRRTGKVPSL 477
Query: 500 SGCFTCHAF 474
F+C +F
Sbjct: 478 --LFSCFSF 484
>gb|AAO15296.1| Unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 426
Score = 43.5 bits (101), Expect = 0.003
Identities = 21/64 (32%), Positives = 35/64 (53%)
Frame = -2
Query: 680 EMKLEKKRASSMDKIMNKLRLAQTKAQEMRSSALANQAHQVTRTSHKVISFRRTGQMGSL 501
E K+E KRA D++ +KL A+ KA+ R +A + + + RT + R+TG + S
Sbjct: 359 EAKIEIKRAREQDRLSSKLAAARHKAEARREAAESRKNQEAARTEEQAAQIRKTGHIPSS 418
Query: 500 SGCF 489
C+
Sbjct: 419 ISCW 422
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 509,579,885
Number of Sequences: 1393205
Number of extensions: 10212060
Number of successful extensions: 19044
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 18675
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19043
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 30270070164
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)