Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002583A_C01 KMC002583A_c01
(856 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
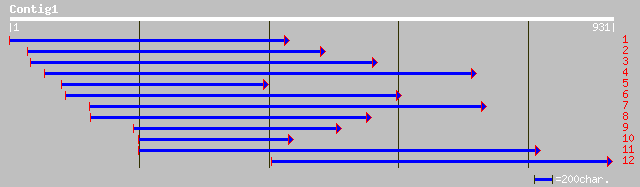
ref|NP_174272.2| unknown protein; protein id: At1g29790.1, suppo... 190 3e-47
gb|AAG46168.1|AC018727_20 hypothetical protein [Oryza sativa] 130 2e-29
ref|NP_198899.1| putative protein; protein id: At5g40830.1 [Arab... 93 4e-18
gb|AAO14627.1|AF467900_4 hypothetical protein [Prunus persica] 93 4e-18
ref|NP_187190.1| hypothetical protein; protein id: At3g05390.1 [... 92 8e-18
>ref|NP_174272.2| unknown protein; protein id: At1g29790.1, supported by cDNA:
gi_20260609 [Arabidopsis thaliana]
gi|20260610|gb|AAM13203.1| unknown protein [Arabidopsis
thaliana]
Length = 378
Score = 190 bits (482), Expect = 3e-47
Identities = 88/111 (79%), Positives = 98/111 (88%)
Frame = -2
Query: 855 VPLHVPLQQRLPLFDGVVDVIRCGRAVNRWIPATMMEFLLFDLDRVLRGGGYLWLDHFFS 676
VPLHVPLQQRLP+FDGVVD++RCGRAVNRWIP T+MEF FDLDR+LRGGGYLWLD FFS
Sbjct: 271 VPLHVPLQQRLPVFDGVVDLVRCGRAVNRWIPVTVMEFFFFDLDRILRGGGYLWLDRFFS 330
Query: 675 KGVDLEKVYAPLIGKLGYKKVKWATGNKTDAGGVKHGEVYLTVLLQKPLSR 523
K VDLE VYAP+IGKLGYKKVKWA NK D+ KHGEV+LT LLQKP++R
Sbjct: 331 KKVDLENVYAPMIGKLGYKKVKWAVANKADS---KHGEVFLTALLQKPVAR 378
>gb|AAG46168.1|AC018727_20 hypothetical protein [Oryza sativa]
Length = 396
Score = 130 bits (328), Expect = 2e-29
Identities = 62/113 (54%), Positives = 76/113 (66%), Gaps = 5/113 (4%)
Frame = -2
Query: 855 VPLHVPLQQRLPLFDGVVDVIRCGRAVNRWIPATMMEFLLFDLDRVLRGGGYLWLDHFFS 676
VPLH PLQQR P+ D +D++R G AVNRWIP +EFL +D DRVLR G LW+DHF+
Sbjct: 284 VPLHAPLQQRFPVGDATMDLVRAGHAVNRWIPEAALEFLWYDADRVLRPRGLLWVDHFWC 343
Query: 675 KGVDLEKVYAPLIGKLGYKKVKWATGNKTD-----AGGVKHGEVYLTVLLQKP 532
+ DL VY P++ +LGYK +KWA +KT G KH VYLT LLQKP
Sbjct: 344 RRPDLAAVYQPMLRRLGYKTLKWAVADKTTPTPTAPPGAKHDHVYLTALLQKP 396
>ref|NP_198899.1| putative protein; protein id: At5g40830.1 [Arabidopsis thaliana]
gi|10177962|dbj|BAB11345.1|
emb|CAB80933.1~gene_id:MHK7.6~similar to unknown protein
[Arabidopsis thaliana] gi|27311741|gb|AAO00836.1|
putative protein [Arabidopsis thaliana]
Length = 414
Score = 93.2 bits (230), Expect = 4e-18
Identities = 47/109 (43%), Positives = 67/109 (61%), Gaps = 2/109 (1%)
Frame = -2
Query: 852 PLHVPLQQRLPLFDGVVDVIRCGRAVNRWIP--ATMMEFLLFDLDRVLRGGGYLWLDHFF 679
PL + L QRLP +D V D+I ++ + +EFL+FDLDR+L+ GG WLD+F+
Sbjct: 309 PLFMSLDQRLPFYDNVFDLIHASNGLDLAVSNKPEKLEFLMFDLDRILKPGGLFWLDNFY 368
Query: 678 SKGVDLEKVYAPLIGKLGYKKVKWATGNKTDAGGVKHGEVYLTVLLQKP 532
+ ++V LI + GYKK+KW G KTDA EV+L+ +LQKP
Sbjct: 369 CGNDEKKRVLTRLIERFGYKKLKWVVGEKTDA------EVFLSAVLQKP 411
>gb|AAO14627.1|AF467900_4 hypothetical protein [Prunus persica]
Length = 421
Score = 93.2 bits (230), Expect = 4e-18
Identities = 42/108 (38%), Positives = 63/108 (57%)
Frame = -2
Query: 852 PLHVPLQQRLPLFDGVVDVIRCGRAVNRWIPATMMEFLLFDLDRVLRGGGYLWLDHFFSK 673
PL + L R P +D V D++ ++ EF++FD+DR+LR GG WLD+F+
Sbjct: 312 PLFLSLDHRFPFYDNVFDLVHAASGLDVGGKPEKFEFVMFDIDRILRPGGLFWLDNFYCS 371
Query: 672 GVDLEKVYAPLIGKLGYKKVKWATGNKTDAGGVKHGEVYLTVLLQKPL 529
+ ++ LI + GYKK+KW G+K DA EVYL+ +LQKP+
Sbjct: 372 NEEKKRDLTRLIERFGYKKLKWVVGDKVDAAASGKSEVYLSAVLQKPV 419
>ref|NP_187190.1| hypothetical protein; protein id: At3g05390.1 [Arabidopsis
thaliana] gi|7596772|gb|AAF64543.1| hypothetical protein
[Arabidopsis thaliana]
Length = 463
Score = 92.4 bits (228), Expect = 8e-18
Identities = 48/108 (44%), Positives = 66/108 (60%)
Frame = -2
Query: 855 VPLHVPLQQRLPLFDGVVDVIRCGRAVNRWIPATMMEFLLFDLDRVLRGGGYLWLDHFFS 676
+PL++ L QRLP FD +D+I ++ WI +M+F+L+D DRVLR GG LW+D FF
Sbjct: 359 IPLYISLNQRLPFFDNTMDMIHTTGLMDGWIDLLLMDFVLYDWDRVLRPGGLLWIDRFFC 418
Query: 675 KGVDLEKVYAPLIGKLGYKKVKWATGNKTDAGGVKHGEVYLTVLLQKP 532
K DL+ Y + + YKK KWA K+ EVYL+ LL+KP
Sbjct: 419 KKKDLDD-YMYMFLQFRYKKHKWAISPKS------KDEVYLSALLEKP 459
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 822,817,597
Number of Sequences: 1393205
Number of extensions: 20817966
Number of successful extensions: 79260
Number of sequences better than 10.0: 334
Number of HSP's better than 10.0 without gapping: 66994
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 77277
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 45152354394
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)