Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002561A_C01 KMC002561A_c01
(948 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
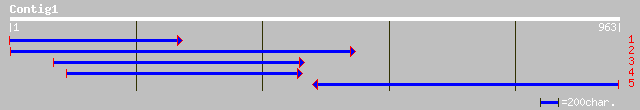
gb|AAO14626.1|AF467900_3 hypothetical protein [Prunus persica] 327 3e-92
ref|NP_198775.1| putative protein; protein id: At5g39590.1, supp... 301 5e-83
gb|AAK74025.1| AT5g39590/MIJ24_60 [Arabidopsis thaliana] gi|2902... 301 5e-83
ref|XP_226525.1| similar to RIKEN cDNA 4632415K11 gene [Mus musc... 48 2e-04
ref|NP_788020.1| CG4658-PA [Drosophila melanogaster] gi|28574111... 45 2e-04
>gb|AAO14626.1|AF467900_3 hypothetical protein [Prunus persica]
Length = 539
Score = 327 bits (839), Expect(2) = 3e-92
Identities = 151/205 (73%), Positives = 178/205 (86%)
Frame = -3
Query: 883 QRSIANLVTASSGDADESHESSSTNRKWVLGALTDQGLENKDIFYGNSGCLYSISPVFHA 704
Q + L +A+SGDA + + RKW +GALT+QG ENKD+FYG+SG LY+ISPVFH
Sbjct: 338 QGPLLMLFSATSGDASDGR---ANERKWTVGALTNQGFENKDLFYGSSGNLYAISPVFHV 394
Query: 703 FPPSGKEKNFVYSHLHPTGKGYQPHPKPAGVAFGGTPGNERIFMDDDFSKLTVRHHAVDK 524
+PP+GKEKNFVYSHLHPTG+ Y+P PKP G+ FGG+ GNERIF+D+DFSK+T+RHHA DK
Sbjct: 395 YPPTGKEKNFVYSHLHPTGRTYEPKPKPVGIGFGGSLGNERIFIDEDFSKVTIRHHAADK 454
Query: 523 TYQSGSLIPDQGFLPVEALISEVEVWGLGGNAAKDVQNSYKRREELFTEQRRKIDLKTFA 344
TYQ GSL PDQGFLPVEALISEVEVWGLGG +AKDVQ+SYK+RE+LFT+QRRK+DLKTFA
Sbjct: 455 TYQPGSLFPDQGFLPVEALISEVEVWGLGGRSAKDVQDSYKKREQLFTDQRRKVDLKTFA 514
Query: 343 NWEDSPEKMMMDMVSDPNAARREER 269
NWEDSPEKMMMDMVSDPNA RRE+R
Sbjct: 515 NWEDSPEKMMMDMVSDPNAVRREDR 539
Score = 34.3 bits (77), Expect(2) = 3e-92
Identities = 12/16 (75%), Positives = 16/16 (100%)
Frame = -1
Query: 912 HRFWSNVEGYKGPLLI 865
+RFWSN+EGY+GPLL+
Sbjct: 328 NRFWSNIEGYQGPLLM 343
>ref|NP_198775.1| putative protein; protein id: At5g39590.1, supported by cDNA:
gi_15010731 [Arabidopsis thaliana]
gi|9758334|dbj|BAB08890.1| gene_id:MIJ24.8~unknown
protein [Arabidopsis thaliana]
Length = 542
Score = 301 bits (772), Expect(2) = 5e-83
Identities = 136/201 (67%), Positives = 164/201 (80%)
Frame = -3
Query: 871 ANLVTASSGDADESHESSSTNRKWVLGALTDQGLENKDIFYGNSGCLYSISPVFHAFPPS 692
A ++ S HE++S+ RKWV+GA+ QG EN+D FYG+SG L+SISPVFHAF S
Sbjct: 342 APILVIISASCKVEHEATSSERKWVIGAILQQGFENRDAFYGSSGNLFSISPVFHAFSSS 401
Query: 691 GKEKNFVYSHLHPTGKGYQPHPKPAGVAFGGTPGNERIFMDDDFSKLTVRHHAVDKTYQS 512
GKEKNF YSHLHP G Y HPKP G+ FGGT GNERIF+D+DF+K+TVRHHAVDKTYQS
Sbjct: 402 GKEKNFAYSHLHPAGGVYDAHPKPVGIGFGGTLGNERIFIDEDFAKITVRHHAVDKTYQS 461
Query: 511 GSLIPDQGFLPVEALISEVEVWGLGGNAAKDVQNSYKRREELFTEQRRKIDLKTFANWED 332
GSL P+QG+LPVEAL+ ++E WGLGGN A+++Q Y++REELFT QRRKIDLKTF NWED
Sbjct: 462 GSLFPNQGYLPVEALVLDIEAWGLGGNKAREIQQKYQKREELFTNQRRKIDLKTFTNWED 521
Query: 331 SPEKMMMDMVSDPNAARREER 269
SPEKMMMDM+ +PNA R+EER
Sbjct: 522 SPEKMMMDMMGNPNAPRKEER 542
Score = 29.6 bits (65), Expect(2) = 5e-83
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = -1
Query: 921 KRHHRFWSNVEGYKGPLLI 865
K +R WSNVEGY P+L+
Sbjct: 328 KGMNRLWSNVEGYHAPILV 346
>gb|AAK74025.1| AT5g39590/MIJ24_60 [Arabidopsis thaliana]
gi|29028740|gb|AAO64749.1| At5g39590/MIJ24_60
[Arabidopsis thaliana]
Length = 541
Score = 301 bits (772), Expect(2) = 5e-83
Identities = 136/201 (67%), Positives = 164/201 (80%)
Frame = -3
Query: 871 ANLVTASSGDADESHESSSTNRKWVLGALTDQGLENKDIFYGNSGCLYSISPVFHAFPPS 692
A ++ S HE++S+ RKWV+GA+ QG EN+D FYG+SG L+SISPVFHAF S
Sbjct: 341 APILVIISASCKVEHEATSSERKWVIGAILQQGFENRDAFYGSSGNLFSISPVFHAFSSS 400
Query: 691 GKEKNFVYSHLHPTGKGYQPHPKPAGVAFGGTPGNERIFMDDDFSKLTVRHHAVDKTYQS 512
GKEKNF YSHLHP G Y HPKP G+ FGGT GNERIF+D+DF+K+TVRHHAVDKTYQS
Sbjct: 401 GKEKNFAYSHLHPAGGVYDAHPKPVGIGFGGTLGNERIFIDEDFAKITVRHHAVDKTYQS 460
Query: 511 GSLIPDQGFLPVEALISEVEVWGLGGNAAKDVQNSYKRREELFTEQRRKIDLKTFANWED 332
GSL P+QG+LPVEAL+ ++E WGLGGN A+++Q Y++REELFT QRRKIDLKTF NWED
Sbjct: 461 GSLFPNQGYLPVEALVLDIEAWGLGGNKAREIQQKYQKREELFTNQRRKIDLKTFTNWED 520
Query: 331 SPEKMMMDMVSDPNAARREER 269
SPEKMMMDM+ +PNA R+EER
Sbjct: 521 SPEKMMMDMMGNPNAPRKEER 541
Score = 29.6 bits (65), Expect(2) = 5e-83
Identities = 11/19 (57%), Positives = 14/19 (72%)
Frame = -1
Query: 921 KRHHRFWSNVEGYKGPLLI 865
K +R WSNVEGY P+L+
Sbjct: 327 KGMNRLWSNVEGYHAPILV 345
>ref|XP_226525.1| similar to RIKEN cDNA 4632415K11 gene [Mus musculus] [Rattus
norvegicus]
Length = 667
Score = 48.1 bits (113), Expect = 2e-04
Identities = 36/129 (27%), Positives = 55/129 (41%), Gaps = 1/129 (0%)
Frame = -3
Query: 802 WVLGALTDQGLENKDIFYGNSGC-LYSISPVFHAFPPSGKEKNFVYSHLHPTGKGYQPHP 626
+V G E K F G++ C L+SI+P + P+G +F+Y + Y
Sbjct: 412 YVFGGFASCSWEVKPQFQGDNKCFLFSIAPRMATYTPTGYNNHFMYLN-------YGQQT 464
Query: 625 KPAGVAFGGTPGNERIFMDDDFSKLTVRHHAVDKTYQSGSLIPDQGFLPVEALISEVEVW 446
P G+ GG +++ DF K + TY S L + F ++EVW
Sbjct: 465 MPNGLGMGGQHHYFGLWVAADFGKGHSKAKPACTTYSSPQLSAQEDF-----QFEKMEVW 519
Query: 445 GLGGNAAKD 419
GLG + KD
Sbjct: 520 GLGNLSEKD 528
>ref|NP_788020.1| CG4658-PA [Drosophila melanogaster] gi|28574111|ref|NP_788021.1|
CG4658-PB [Drosophila melanogaster]
gi|28574113|ref|NP_788022.1| CG4658-PC [Drosophila
melanogaster] gi|7297585|gb|AAF52839.1| CG4658-PA
[Drosophila melanogaster]
gi|22946073|gb|AAN10715.1|AE003626_51 CG4658-PB
[Drosophila melanogaster]
gi|22946074|gb|AAN10716.1|AE003626_52 CG4658-PC
[Drosophila melanogaster] gi|28557627|gb|AAO45219.1|
RE10231p [Drosophila melanogaster]
Length = 526
Score = 44.7 bits (104), Expect(2) = 2e-04
Identities = 37/152 (24%), Positives = 68/152 (44%), Gaps = 1/152 (0%)
Frame = -3
Query: 757 IFYGNSG-CLYSISPVFHAFPPSGKEKNFVYSHLHPTGKGYQPHPKPAGVAFGGTPGNER 581
+F G G C+ + P F K+ N +Y L+ + +GY P G+ G P
Sbjct: 398 LFVGGEGSCVIQLLPKFVILE---KKPNILY--LNTSIRGY-----PKGLRAGADPRKPI 447
Query: 580 IFMDDDFSKLTVRHHAVDKTYQSGSLIPDQGFLPVEALISEVEVWGLGGNAAKDVQNSYK 401
I +D+ F + + A A + +EVWG G ++++VQ K
Sbjct: 448 IAVDEHFENIDCKGLA--------------------AGLMSIEVWGCGDKSSREVQLDIK 487
Query: 400 RREELFTEQRRKIDLKTFANWEDSPEKMMMDM 305
+ + E++R + L T A+W D P++ ++++
Sbjct: 488 KWQIKEAERQRTVKL-TAADWMDHPDRYLLEL 518
Score = 22.7 bits (47), Expect(2) = 2e-04
Identities = 9/18 (50%), Positives = 13/18 (72%)
Frame = -1
Query: 912 HRFWSNVEGYKGPLLIWL 859
+RF +V GY+GP L+ L
Sbjct: 360 NRFLHHVLGYRGPTLVLL 377
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 885,624,126
Number of Sequences: 1393205
Number of extensions: 20909778
Number of successful extensions: 50978
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 48424
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 50882
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 53246406144
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)