Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002548A_C01 KMC002548A_c01
(584 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
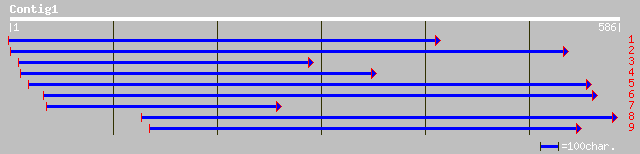
pir||A86366 T26J12.6 protein - Arabidopsis thaliana gi|2829894|g... 55 5e-07
ref|NP_173730.1| unknown protein; protein id: At1g23170.1 [Arabi... 55 5e-07
dbj|BAB03379.1| unnamed protein product [Oryza sativa (japonica ... 53 2e-06
ref|NP_177234.1| unknown protein; protein id: At1g70770.1 [Arabi... 49 5e-05
gb|AAF23204.1|AC016795_17 unknown protein [Arabidopsis thaliana] 39 0.037
>pir||A86366 T26J12.6 protein - Arabidopsis thaliana gi|2829894|gb|AAC00602.1|
Unknown protein [Arabidopsis thaliana]
Length = 1299
Score = 55.5 bits (132), Expect = 5e-07
Identities = 31/63 (49%), Positives = 41/63 (64%), Gaps = 4/63 (6%)
Frame = -1
Query: 584 KHENALTKVDDGARHGLLKDSDKYCKVILGRLSRGNGCMKSMAFVSVVLA----VGAVFM 417
K+E ALT+ +G L KD+DKYCKVI G+LS G GC+KS+A + +LA GA +
Sbjct: 409 KNEEALTEGGNGVSQSLYKDADKYCKVIAGKLSSG-GCIKSIATTAALLAATGFAGAAAL 467
Query: 416 SQN 408
S N
Sbjct: 468 SAN 470
>ref|NP_173730.1| unknown protein; protein id: At1g23170.1 [Arabidopsis thaliana]
Length = 569
Score = 55.5 bits (132), Expect = 5e-07
Identities = 31/63 (49%), Positives = 41/63 (64%), Gaps = 4/63 (6%)
Frame = -1
Query: 584 KHENALTKVDDGARHGLLKDSDKYCKVILGRLSRGNGCMKSMAFVSVVLA----VGAVFM 417
K+E ALT+ +G L KD+DKYCKVI G+LS G GC+KS+A + +LA GA +
Sbjct: 479 KNEEALTEGGNGVSQSLYKDADKYCKVIAGKLSSG-GCIKSIATTAALLAATGFAGAAAL 537
Query: 416 SQN 408
S N
Sbjct: 538 SAN 540
>dbj|BAB03379.1| unnamed protein product [Oryza sativa (japonica cultivar-group)]
Length = 586
Score = 53.1 bits (126), Expect = 2e-06
Identities = 32/74 (43%), Positives = 44/74 (59%), Gaps = 2/74 (2%)
Frame = -1
Query: 584 KHENALTKVDDGARHGLLKDSDKYCKVILGRLSRGNGCMKSMAFVSVVLAVGAVF-MSQN 408
K+E AL D + +K +DKY K ILGRLSRG C+K V + LAV A F +S N
Sbjct: 508 KNEAALEAATDSGKQASIKAADKYSKEILGRLSRGGACLKGSLLV-ITLAVAAGFVLSPN 566
Query: 407 MHL-WDYDKISEML 369
+ + D+DK+ M+
Sbjct: 567 LEIPSDWDKLQAMV 580
>ref|NP_177234.1| unknown protein; protein id: At1g70770.1 [Arabidopsis thaliana]
gi|25406072|pir||C96732 hypothetical protein F15H11.2
[imported] - Arabidopsis thaliana
gi|5902390|gb|AAD55492.1|AC008148_2 Unknown protein
[Arabidopsis thaliana]
gi|12324751|gb|AAG52333.1|AC011663_12 unknown protein;
13405-15968 [Arabidopsis thaliana]
gi|22531184|gb|AAM97096.1| unknown protein [Arabidopsis
thaliana]
Length = 610
Score = 48.9 bits (115), Expect = 5e-05
Identities = 25/50 (50%), Positives = 32/50 (64%)
Frame = -1
Query: 584 KHENALTKVDDGARHGLLKDSDKYCKVILGRLSRGNGCMKSMAFVSVVLA 435
K+E +T + GA L K++DK CK I GRLSRG+GC+K A V LA
Sbjct: 517 KNEEVIT--EGGANASLYKEADKSCKTISGRLSRGSGCLKGTAITLVFLA 564
>gb|AAF23204.1|AC016795_17 unknown protein [Arabidopsis thaliana]
Length = 459
Score = 39.3 bits (90), Expect = 0.037
Identities = 19/60 (31%), Positives = 32/60 (52%)
Frame = -1
Query: 584 KHENALTKVDDGARHGLLKDSDKYCKVILGRLSRGNGCMKSMAFVSVVLAVGAVFMSQNM 405
K+E A+T + G K++DK CKV+ GR N C+K A ++ V+ ++ N+
Sbjct: 402 KNEKAIT--EGGTIGSRYKEADKSCKVMSGRFFPENACLKGTAIITAVVLAAVAVLASNL 459
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 463,765,972
Number of Sequences: 1393205
Number of extensions: 9266906
Number of successful extensions: 23539
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 21739
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23533
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)