Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002544A_C01 KMC002544A_c01
(586 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
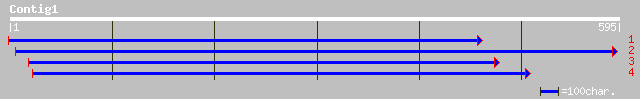
emb|CAA04664.1| hypothetical protein [Citrus x paradisi] 218 3e-56
ref|NP_189345.1| gda-1, putative; protein id: At3g27090.1, suppo... 207 8e-53
gb|AAM64572.1| gda-1, putative [Arabidopsis thaliana] 207 1e-52
emb|CAD37200.1| GDA2 protein [Pisum sativum] 152 3e-36
ref|NP_568600.1| putative protein; protein id: At5g42050.1, supp... 83 3e-15
>emb|CAA04664.1| hypothetical protein [Citrus x paradisi]
Length = 305
Score = 218 bits (556), Expect = 3e-56
Identities = 105/175 (60%), Positives = 141/175 (80%), Gaps = 5/175 (2%)
Frame = +3
Query: 75 MGDELRGHTKVSEDDHKWFMVASKLAEQTRLKGERLNNLDLSKGPIETRPREKMGFQEEN 254
+GDELRG ++ SED W AS+LAEQTR KGER+NNLDLSKG E RPR+K+ + E+N
Sbjct: 10 LGDELRGQSRTSEDQ-SWLRAASRLAEQTRFKGERMNNLDLSKGMTEIRPRDKIMYHEDN 68
Query: 255 KFDTLNFS--MLNLESRISENMSKSSFRNGVYNMNAVYQKSNANLVGNININKYNGN--- 419
F++ NF+ M+NL++++ EN++KSS RNG+YNMNAVYQK++ + +GN+ +NKY GN
Sbjct: 69 NFESFNFNFNMMNLDNKVVENVTKSSLRNGIYNMNAVYQKNSGHNMGNLMVNKYGGNNLS 128
Query: 420 VQLNKDSNNISNNNNESNSSNVAEKRFKTLPAAETLPRNEVLGGYICVCNNDTMQ 584
V+ +++NN +NNNN+SN+++ +KRFKTLPA ETLPRNEVLGGYI VCNNDTMQ
Sbjct: 129 VKEAENNNNNNNNNNDSNANSALDKRFKTLPATETLPRNEVLGGYIFVCNNDTMQ 183
>ref|NP_189345.1| gda-1, putative; protein id: At3g27090.1, supported by cDNA: 3049.,
supported by cDNA: gi_16604449 [Arabidopsis thaliana]
gi|9279632|dbj|BAB01090.1| contains similarity to
unknown protein~emb|CAA04664.1~gene_id:MOJ10.16
[Arabidopsis thaliana] gi|16604450|gb|AAL24231.1|
AT3g27090/MOJ10_18 [Arabidopsis thaliana]
gi|21655279|gb|AAM65351.1| AT3g27090/MOJ10_18
[Arabidopsis thaliana]
Length = 296
Score = 207 bits (527), Expect = 8e-53
Identities = 107/174 (61%), Positives = 133/174 (75%), Gaps = 4/174 (2%)
Frame = +3
Query: 75 MGDELRGHTKVSEDDHKWFMVASKLAEQTRLKGERLNNLDLSKGPIETRPREKMGFQEEN 254
+GDELRG T+ SED HKW VA+KLAEQTR+KGER+NNLDLSKG E RP EK FQE N
Sbjct: 7 LGDELRGQTRASED-HKWSTVATKLAEQTRMKGERMNNLDLSKGYTEFRPSEKFSFQENN 65
Query: 255 KFDTLNFSMLNLESRISEN-MSKSSFRNGVYNMNAVYQKSNANLVGNININKYNGNVQLN 431
LNF+MLNL+ + E+ M K+S ++ VYNMN V+QK++ GN+ +NKYNGNV N
Sbjct: 66 ----LNFNMLNLDGKFGESIMGKTSMQSNVYNMNTVFQKNDFKSGGNMKVNKYNGNVVAN 121
Query: 432 KDSNNISNNNNESNSSNV---AEKRFKTLPAAETLPRNEVLGGYICVCNNDTMQ 584
K+ +N +NNN +++ N+ +KRFKTLPA+ETLPRNEVLGGYI VCNNDTMQ
Sbjct: 122 KEMSNNKHNNNCNDNGNMNLAVDKRFKTLPASETLPRNEVLGGYIFVCNNDTMQ 175
>gb|AAM64572.1| gda-1, putative [Arabidopsis thaliana]
Length = 296
Score = 207 bits (526), Expect = 1e-52
Identities = 106/174 (60%), Positives = 133/174 (75%), Gaps = 4/174 (2%)
Frame = +3
Query: 75 MGDELRGHTKVSEDDHKWFMVASKLAEQTRLKGERLNNLDLSKGPIETRPREKMGFQEEN 254
+GDELRG T+ SED HKW VA+KLAEQTR+KGER+NNLDLSKG E RP EK FQE N
Sbjct: 7 LGDELRGQTRASED-HKWSTVATKLAEQTRMKGERMNNLDLSKGYTEFRPSEKFSFQENN 65
Query: 255 KFDTLNFSMLNLESRISEN-MSKSSFRNGVYNMNAVYQKSNANLVGNININKYNGNVQLN 431
LNF+MLNL+ + E+ M K+S ++ VYNMN V+QK++ GN+ +NKYNGN+ N
Sbjct: 66 ----LNFNMLNLDGKFGESIMGKTSMQSNVYNMNTVFQKNDFKSGGNMKVNKYNGNIVAN 121
Query: 432 KDSNNISNNNNESNSSNV---AEKRFKTLPAAETLPRNEVLGGYICVCNNDTMQ 584
K+ +N +NNN +++ N+ +KRFKTLPA+ETLPRNEVLGGYI VCNNDTMQ
Sbjct: 122 KEMSNNKHNNNCNDNGNMNLAVDKRFKTLPASETLPRNEVLGGYIFVCNNDTMQ 175
>emb|CAD37200.1| GDA2 protein [Pisum sativum]
Length = 213
Score = 152 bits (384), Expect = 3e-36
Identities = 71/91 (78%), Positives = 81/91 (88%)
Frame = +3
Query: 312 MSKSSFRNGVYNMNAVYQKSNANLVGNININKYNGNVQLNKDSNNISNNNNESNSSNVAE 491
M+K+S RNGVYNMNAVYQKSNAN VGN+N NKY+GNVQLNKD ++ +NNNN N++N +
Sbjct: 1 MNKNSLRNGVYNMNAVYQKSNANFVGNMNSNKYSGNVQLNKDPHSNNNNNNNENNTNATD 60
Query: 492 KRFKTLPAAETLPRNEVLGGYICVCNNDTMQ 584
KRFKTLPAAETLPRNEVLGGYI VCNNDTMQ
Sbjct: 61 KRFKTLPAAETLPRNEVLGGYIFVCNNDTMQ 91
>ref|NP_568600.1| putative protein; protein id: At5g42050.1, supported by cDNA:
6903., supported by cDNA: gi_19424060, supported by
cDNA: gi_21281017 [Arabidopsis thaliana]
gi|19424061|gb|AAL87257.1| unknown protein [Arabidopsis
thaliana] gi|21281018|gb|AAM45105.1| unknown protein
[Arabidopsis thaliana] gi|21594375|gb|AAM66001.1|
unknown [Arabidopsis thaliana]
Length = 349
Score = 82.8 bits (203), Expect = 3e-15
Identities = 53/139 (38%), Positives = 71/139 (50%), Gaps = 10/139 (7%)
Frame = +3
Query: 198 SKGPIETRPREKMG--FQEENKFDTLNFSMLNLESRISENMSKSSFRNGVYNMNAVYQKS 371
SK ++ P +K F + KF+++N +N+ + F GVY Y
Sbjct: 96 SKSTVDLNPIDKFNSPFNDTWKFNSVN---VNVNGYSPSSAVNGDFNKGVYTSMKKYGY- 151
Query: 372 NANLVGNININK--------YNGNVQLNKDSNNISNNNNESNSSNVAEKRFKTLPAAETL 527
N NL N N NK G + K+ N +N NE + +N +KRFKTLP AE L
Sbjct: 152 NVNLKNN-NKNKGIDEDHQIQKGGKKNRKNQQNNNNQRNEDDKNNGLDKRFKTLPPAEAL 210
Query: 528 PRNEVLGGYICVCNNDTMQ 584
PRNE +GGYI VCNNDTM+
Sbjct: 211 PRNETIGGYIFVCNNDTME 229
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 515,312,796
Number of Sequences: 1393205
Number of extensions: 11293477
Number of successful extensions: 169816
Number of sequences better than 10.0: 2760
Number of HSP's better than 10.0 without gapping: 45510
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 87484
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21997688174
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)