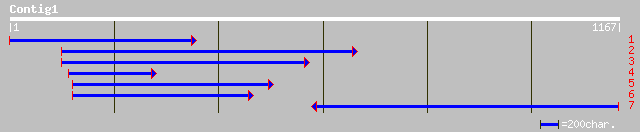
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002529A_C01 KMC002529A_c01
(1161 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S51529 SPF1 protein - sweet potato gi|484261|dbj|BAA06278.1... 298 8e-80
dbj|BAA77383.1| transcription factor NtWRKY2 [Nicotiana tabacum] 290 4e-77
dbj|BAB61055.1| WRKY DNA-binding protein [Nicotiana tabacum] 265 1e-69
gb|AAC31956.1| zinc finger protein; WRKY1 [Pimpinella brachycarpa] 260 3e-68
pir||S61413 DNA-binding protein ABF1 - wild oat (fragment) gi|11... 260 3e-68
>pir||S51529 SPF1 protein - sweet potato gi|484261|dbj|BAA06278.1| SPF1 protein
[Ipomoea batatas]
Length = 549
Score = 298 bits (764), Expect = 8e-80
Identities = 162/237 (68%), Positives = 182/237 (76%), Gaps = 4/237 (1%)
Frame = -2
Query: 1160 FEQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDD 981
FEQSSQK SGGDE DEDEPD+KRWKVE E+EG+SA GSRTVREPRVVVQTTSDIDILDD
Sbjct: 328 FEQSSQKRESGGDEFDEDEPDAKRWKVENESEGVSAQGSRTVREPRVVVQTTSDIDILDD 387
Query: 980 GYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAA 801
GYRWRKYGQKVVKGNPNPRSYYKCT GCPVRKHVERASHD+R+VITTYEGKHNHDVPAA
Sbjct: 388 GYRWRKYGQKVVKGNPNPRSYYKCTSQGCPVRKHVERASHDIRSVITTYEGKHNHDVPAA 447
Query: 800 RGSGNHSVTRPL-PNNT---ANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFT 633
RGSG+H + R PNN A AIRPS+++ Q ++ + S RP Q + +P+
Sbjct: 448 RGSGSHGLNRGANPNNNAAMAMAIRPSTMSLQSNYPI---PIPSTRPMQQGE--GQAPY- 501
Query: 632 LEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPRDDTFFDSLL 462
EML Q GGFG+ SGFGN M +Y Q+ DN F SRAKEEPRDD F D+LL
Sbjct: 502 -EML-QGSGGFGY-SGFGNPMNAY------ANQIQDNAF-SRAKEEPRDDLFLDTLL 548
Score = 95.1 bits (235), Expect = 2e-18
Identities = 58/156 (37%), Positives = 75/156 (47%), Gaps = 3/156 (1%)
Frame = -2
Query: 1052 PGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVE 873
P S+T+RE R DDGY WRKYGQK VKG+ NPRSYYKCTHP CP +K VE
Sbjct: 198 PPSQTLREQRRS----------DDGYNWRKYGQKQVKGSENPRSYYKCTHPNCPTKKKVE 247
Query: 872 RASHDLRAVITTYEGKHNHDVPAARGSGNHSVTRPLPNNTANAIR--PSSVTHQHHHNTN 699
RA D + Y+G HNH P + + S A + S V Q + +
Sbjct: 248 RAL-DGQITEIVYKGAHNHPKPQSTRRSSSSTASSASTLAAQSYNAPASDVPDQSYWSNG 306
Query: 698 NNSLQSL-RPQQAPDQVQSSPFTLEMLQQNQGGFGF 594
N + S+ P+ + V F ++ GG F
Sbjct: 307 NGQMDSVATPENSSISVGDDEFEQSSQKRESGGDEF 342
>dbj|BAA77383.1| transcription factor NtWRKY2 [Nicotiana tabacum]
Length = 353
Score = 290 bits (741), Expect = 4e-77
Identities = 160/240 (66%), Positives = 177/240 (73%), Gaps = 7/240 (2%)
Frame = -2
Query: 1157 EQSSQKCRSGGDELDEDEPDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDIDILDDG 978
EQSSQK RS GD+ +E+EPDSKRWK E E+EG+SAPGSRTVREPRVVVQTTSDIDILDDG
Sbjct: 139 EQSSQKSRSRGDDNEEEEPDSKRWKRESESEGLSAPGSRTVREPRVVVQTTSDIDILDDG 198
Query: 977 YRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAAR 798
YRWRKYGQKVVKGNPNPR YYKCT PGCPVRKHVERAS D+R+VITTYEGKHNHDVPAAR
Sbjct: 199 YRWRKYGQKVVKGNPNPRGYYKCTSPGCPVRKHVERASQDIRSVITTYEGKHNHDVPAAR 258
Query: 797 GSGNHSVTRPL-PNNTAN------AIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSP 639
GSG + RP+ PN T N AIRPS T+ LQS+RPQQ SP
Sbjct: 259 GSG---INRPVAPNITYNNGANAMAIRPSV--------TSQIPLQSIRPQQ-------SP 300
Query: 638 FTLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPRDDTFFDSLLC 459
FTLEML + GF SG+ NS SY + QL DN SRAK+EPRDD F ++LLC
Sbjct: 301 FTLEMLHKPSNYNGF-SGYVNSEDSY------ENQLQDNNVFSRAKDEPRDDMFMETLLC 353
Score = 84.7 bits (208), Expect = 2e-15
Identities = 42/76 (55%), Positives = 49/76 (64%), Gaps = 3/76 (3%)
Frame = -2
Query: 986 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 807
+DGY WRKYGQK VKG+ NPRSYYKCT P CP +K VER S D + Y+G HNH P
Sbjct: 28 EDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVER-SLDGQITEIVYKGNHNHPKP 86
Query: 806 AA---RGSGNHSVTRP 768
+ S S+TRP
Sbjct: 87 QSTRRSSSTASSLTRP 102
>dbj|BAB61055.1| WRKY DNA-binding protein [Nicotiana tabacum]
Length = 378
Score = 265 bits (677), Expect = 1e-69
Identities = 147/240 (61%), Positives = 163/240 (67%), Gaps = 7/240 (2%)
Frame = -2
Query: 1157 EQSSQKCRSGGDELDEDE------PDSKRWKVEGENEGISAPGSRTVREPRVVVQTTSDI 996
+ +K RS GD+ DE+E PD KRWK E E+EG+SAPGSRTVREPRVVVQTTSDI
Sbjct: 165 DDDHEKSRSRGDDFDEEEEPDSKEPDPKRWKRESESEGLSAPGSRTVREPRVVVQTTSDI 224
Query: 995 DILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNH 816
DILDDGYRWRKYGQKVVKGNPNPRSYYKCT PGCPVRKHVERAS D+R+VITTYEGKHNH
Sbjct: 225 DILDDGYRWRKYGQKVVKGNPNPRSYYKCTSPGCPVRKHVERASQDIRSVITTYEGKHNH 284
Query: 815 DVPAARGSG-NHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSP 639
DVPAARGS N V + N A IRPS + Q PQQ SP
Sbjct: 285 DVPAARGSAINRPVAPTITYNNAIPIRPSVTS------------QIPLPQQ-------SP 325
Query: 638 FTLEMLQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPRDDTFFDSLLC 459
FTLEML + GF SG+ S SY + QL DN SRAK+EPRDD F ++LLC
Sbjct: 326 FTLEMLHKPSNYNGF-SGYATSEDSY------ENQLQDNNVFSRAKDEPRDDMFMETLLC 378
Score = 84.7 bits (208), Expect = 2e-15
Identities = 46/135 (34%), Positives = 68/135 (50%), Gaps = 8/135 (5%)
Frame = -2
Query: 986 DDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVP 807
+DGY WRKYGQK VKG+ NPRSYYKCT P CP +K VER D + Y+G HNH P
Sbjct: 69 EDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERCL-DGQITEIVYKGNHNHPKP 127
Query: 806 --AARGSGNHSV------TRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQV 651
+ R S + ++ T +P++ + S H + + ++ PD
Sbjct: 128 TQSTRRSSSLAIQPYNTQTNEIPDHQSTPENSSISFGDDDHEKSRSRGDDFDEEEEPDSK 187
Query: 650 QSSPFTLEMLQQNQG 606
+ P + +++G
Sbjct: 188 EPDPKRWKRESESEG 202
>gb|AAC31956.1| zinc finger protein; WRKY1 [Pimpinella brachycarpa]
Length = 515
Score = 260 bits (665), Expect = 3e-68
Identities = 148/234 (63%), Positives = 173/234 (73%), Gaps = 1/234 (0%)
Frame = -2
Query: 1160 FEQSSQKCRSGGDELDEDEPDSKRWKVEGEN-EGISAPGSRTVREPRVVVQTTSDIDILD 984
FEQ S + G D DE+EPD+KRWK E EN E +S+ GSRTVREPR+VVQTTSDIDILD
Sbjct: 297 FEQGSMN-KQGDD--DENEPDAKRWKGEYENNETMSSLGSRTVREPRIVVQTTSDIDILD 353
Query: 983 DGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPA 804
DGYRWRKYGQKVVKGNPNPRSYYKCT GCPVRKHVERASHDLRAVITTYEGKHNHDVPA
Sbjct: 354 DGYRWRKYGQKVVKGNPNPRSYYKCTQVGCPVRKHVERASHDLRAVITTYEGKHNHDVPA 413
Query: 803 ARGSGNHSVTRPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLEM 624
RGSG+++V RP +NTA P+++ ++ N LQ+ R Q A Q +PFTLEM
Sbjct: 414 PRGSGSYAVNRP-SDNTATTSAPTAIRPTTNY---LNPLQNTRAQPANGQ---APFTLEM 466
Query: 623 LQQNQGGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPRDDTFFDSLL 462
LQ+ + + F SGFGNS +Y +N QQ FS+ AK+EP D+FFDS L
Sbjct: 467 LQRPR-SYEF-SGFGNSTNTYTINQNQQ---ASGQFST-AKDEPDVDSFFDSFL 514
Score = 85.1 bits (209), Expect = 2e-15
Identities = 42/85 (49%), Positives = 51/85 (59%), Gaps = 1/85 (1%)
Frame = -2
Query: 1013 QTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTY 834
QT D LDDGY WRKYGQK VKG+ NPRSYYKCT+ CP +K VE + D Y
Sbjct: 182 QTNRDQSKLDDGYNWRKYGQKQVKGSENPRSYYKCTYLNCPTKKKVE-TTFDGHITEIVY 240
Query: 833 EGKHNHDVP-AARGSGNHSVTRPLP 762
+G HNH P + + S + S +P
Sbjct: 241 KGNHNHPKPQSTKRSSSQSYQNSIP 265
>pir||S61413 DNA-binding protein ABF1 - wild oat (fragment)
gi|1159877|emb|CAA88326.1| DNA-binding protein [Avena
fatua]
Length = 402
Score = 260 bits (665), Expect = 3e-68
Identities = 138/226 (61%), Positives = 160/226 (70%), Gaps = 7/226 (3%)
Frame = -2
Query: 1130 GGDELDEDEPDSKRWKVEGENEGI-SAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQ 954
GGD+LD+DEPDSK+W+ +G+ EG S G+RTVREPRVVVQT SDIDILDDGYRWRKYGQ
Sbjct: 176 GGDDLDDDEPDSKKWRKDGDGEGSNSMAGNRTVREPRVVVQTMSDIDILDDGYRWRKYGQ 235
Query: 953 KVVKGNPNPRSYYKCTHPGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSGNHSVT 774
KVVKGNPNPRSYYKCT GCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGS ++
Sbjct: 236 KVVKGNPNPRSYYKCTTVGCPVRKHVERASHDLRAVITTYEGKHNHDVPAARGSA--ALY 293
Query: 773 RPLPNNTANAIRPSSVTHQHHHNTNNNSLQSLRPQQAPDQVQSSPFTLEMLQQNQ----- 609
RP P A++ S + Q+ N S + + A Q +P QNQ
Sbjct: 294 RPAPRAAADS--AMSTSQQYTANQQQPSAMTYQTSAAAGTQQYAPRPDGFGSQNQGSFGF 351
Query: 608 -GGFGFSSGFGNSMGSYNMNNQQQQQLPDNVFSSRAKEEPRDDTFF 474
G FGFS+GF N GSY +QQQQ+ D + +S AKEEPR+D FF
Sbjct: 352 NGSFGFSAGFDNPTGSYMSQHQQQQRQNDAMQASGAKEEPREDMFF 397
Score = 85.1 bits (209), Expect = 2e-15
Identities = 46/93 (49%), Positives = 53/93 (56%), Gaps = 2/93 (2%)
Frame = -2
Query: 1061 ISAPGSRTVREPRVVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTHPGCPVRK 882
+ A S V QT S DDGY WRKYGQK VKG+ NPRSYYKCT P CP +K
Sbjct: 28 VQATSSEMAPSGGVYRQTHSQRRSSDDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKK 87
Query: 881 HVERASHDLRAVITTYEGKHNHDVPAA--RGSG 789
VE S + + Y+G HNH P + RGSG
Sbjct: 88 KVE-TSIEGQITEIVYKGTHNHAKPLSTRRGSG 119
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,064,292,034
Number of Sequences: 1393205
Number of extensions: 25712294
Number of successful extensions: 82925
Number of sequences better than 10.0: 282
Number of HSP's better than 10.0 without gapping: 70258
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 80699
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 71654580342
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)