Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002526A_C01 KMC002526A_c01
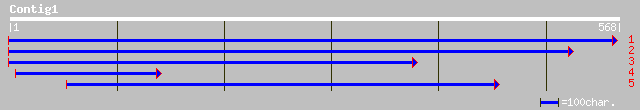
(567 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAD09518.1| NTGP4 [Nicotiana tabacum] 100 1e-20
gb|AAN60299.1| unknown [Arabidopsis thaliana] 95 7e-19
pir||D86463 hypothetical protein F12G12.21 - Arabidopsis thalian... 95 7e-19
gb|AAG52214.1|AC022288_13 AIG1-like protein, 5' partial; 1-1205 ... 95 7e-19
ref|NP_564431.1| expressed protein; protein id: At1g33970.1, sup... 95 7e-19
>gb|AAD09518.1| NTGP4 [Nicotiana tabacum]
Length = 344
Score = 100 bits (249), Expect = 1e-20
Identities = 63/129 (48%), Positives = 86/129 (65%), Gaps = 13/129 (10%)
Frame = -2
Query: 566 SDELFAELKKGAMELHNQQRQVN----LQGQEILEYRKQAKQEYDDQLKRISEMVESKLK 399
+D+LF ELKKGA++L NQ +VN QEILE ++Q ++ Y++QL+RI+E+VESKLK
Sbjct: 220 TDDLFKELKKGAIKLRNQATEVNNLVGYSKQEILELKEQMQKSYEEQLRRITEVVESKLK 279
Query: 398 AATVRLEQQLKEEQAARL-------KAQENANKEIRELREQLFKAEENANKAREELRKHG 240
T RLE+QL +EQAARL +AQ+ ++ EIR+LRE L E A + EELR
Sbjct: 280 DTTHRLEEQLAKEQAARLEAELSAKEAQKKSDNEIRKLREYL----ERAQRETEELRGRS 335
Query: 239 ENR--CAIL 219
+R C IL
Sbjct: 336 ADRGVCNIL 344
>gb|AAN60299.1| unknown [Arabidopsis thaliana]
Length = 337
Score = 94.7 bits (234), Expect = 7e-19
Identities = 55/118 (46%), Positives = 80/118 (67%), Gaps = 11/118 (9%)
Frame = -2
Query: 566 SDELFAELKKGAMELHNQQRQVNL----QGQEILEYRKQAKQEYDDQLKRISEMVESKLK 399
SDELF EL++ A++L +Q+++V L EI E++KQ YD QL RI+EMVE+KL+
Sbjct: 216 SDELFHELQEEAIKLRDQKKEVELLQGYSNNEIDEFKKQIDMSYDRQLSRITEMVETKLR 275
Query: 398 AATVRLEQQLKEEQAARLKAQENANK-------EIRELREQLFKAEENANKAREELRK 246
RLEQQL EEQAARL+A++ AN+ EI++LRE L +AE+ + +++L K
Sbjct: 276 DTAKRLEQQLGEEQAARLEAEKRANEVQKRSSDEIKKLRENLERAEKETKELQKKLGK 333
>pir||D86463 hypothetical protein F12G12.21 - Arabidopsis thaliana
gi|10086478|gb|AAG12538.1|AC015446_19 Similar to AIG1
protein [Arabidopsis thaliana]
gi|10092443|gb|AAG12846.1|AC079286_3 disease resistance
protein AIG1; 5333-4002 [Arabidopsis thaliana]
gi|21593218|gb|AAM65167.1| AIG1-like protein, 5' partial
[Arabidopsis thaliana]
Length = 337
Score = 94.7 bits (234), Expect = 7e-19
Identities = 55/118 (46%), Positives = 80/118 (67%), Gaps = 11/118 (9%)
Frame = -2
Query: 566 SDELFAELKKGAMELHNQQRQVNL----QGQEILEYRKQAKQEYDDQLKRISEMVESKLK 399
SDELF EL++ A++L +Q+++V L EI E++KQ YD QL RI+EMVE+KL+
Sbjct: 216 SDELFHELQEEAIKLRDQKKEVELLQGYSNNEIDEFKKQIDMSYDRQLSRITEMVETKLR 275
Query: 398 AATVRLEQQLKEEQAARLKAQENANK-------EIRELREQLFKAEENANKAREELRK 246
RLEQQL EEQAARL+A++ AN+ EI++LRE L +AE+ + +++L K
Sbjct: 276 DTAKRLEQQLGEEQAARLEAEKRANEVQKRSSDEIKKLRENLERAEKETKELQKKLGK 333
>gb|AAG52214.1|AC022288_13 AIG1-like protein, 5' partial; 1-1205 [Arabidopsis thaliana]
Length = 294
Score = 94.7 bits (234), Expect = 7e-19
Identities = 55/118 (46%), Positives = 80/118 (67%), Gaps = 11/118 (9%)
Frame = -2
Query: 566 SDELFAELKKGAMELHNQQRQVNL----QGQEILEYRKQAKQEYDDQLKRISEMVESKLK 399
SDELF EL++ A++L +Q+++V L EI E++KQ YD QL RI+EMVE+KL+
Sbjct: 173 SDELFHELQEEAIKLRDQKKEVELLQGYSNNEIDEFKKQIDMSYDRQLSRITEMVETKLR 232
Query: 398 AATVRLEQQLKEEQAARLKAQENANK-------EIRELREQLFKAEENANKAREELRK 246
RLEQQL EEQAARL+A++ AN+ EI++LRE L +AE+ + +++L K
Sbjct: 233 DTAKRLEQQLGEEQAARLEAEKRANEVQKRSSDEIKKLRENLERAEKETKELQKKLGK 290
>ref|NP_564431.1| expressed protein; protein id: At1g33970.1, supported by cDNA:
37204. [Arabidopsis thaliana]
Length = 342
Score = 94.7 bits (234), Expect = 7e-19
Identities = 55/118 (46%), Positives = 80/118 (67%), Gaps = 11/118 (9%)
Frame = -2
Query: 566 SDELFAELKKGAMELHNQQRQVNL----QGQEILEYRKQAKQEYDDQLKRISEMVESKLK 399
SDELF EL++ A++L +Q+++V L EI E++KQ YD QL RI+EMVE+KL+
Sbjct: 221 SDELFHELQEEAIKLRDQKKEVELLQGYSNNEIDEFKKQIDMSYDRQLSRITEMVETKLR 280
Query: 398 AATVRLEQQLKEEQAARLKAQENANK-------EIRELREQLFKAEENANKAREELRK 246
RLEQQL EEQAARL+A++ AN+ EI++LRE L +AE+ + +++L K
Sbjct: 281 DTAKRLEQQLGEEQAARLEAEKRANEVQKRSSDEIKKLRENLERAEKETKELQKKLGK 338
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 421,102,054
Number of Sequences: 1393205
Number of extensions: 8167206
Number of successful extensions: 37751
Number of sequences better than 10.0: 1639
Number of HSP's better than 10.0 without gapping: 32883
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36894
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20669577624
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)