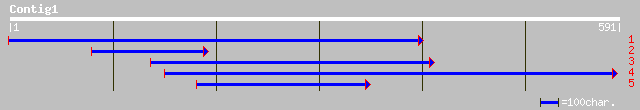
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002520A_C01 KMC002520A_c01
(506 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_006816.2| transmembrane protein (63kD), endoplasmic retic... 41 0.007
pir||S33377 P63 protein - human 41 0.007
emb|CAA65506.1| cuticular collagen [Teladorsagia circumcincta] 40 0.020
emb|CAA65507.1| cuticular collagen [Teladorsagia circumcincta] 38 0.076
ref|NP_037279.1| chloride channel 1; Chloride channel 1 (skeleta... 37 0.099
>ref|NP_006816.2| transmembrane protein (63kD), endoplasmic reticulum/Golgi interm;
transmembrane protein (63kD), endoplasmic
reticulum/Golgi intermediate compartment [Homo sapiens]
gi|19572637|emb|CAA49535.2| P63 protein [Homo sapiens]
Length = 602
Score = 41.2 bits (95), Expect = 0.007
Identities = 27/70 (38%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Frame = -2
Query: 421 AGAPFEQVADPRDAKGVLASQPMHLP--PPPPPLPH-MSHLKQ---NYYSGEPGHDGQGY 260
A +P E+ A P +A +P P PPPPP PH H +Q N G+ GH G G
Sbjct: 15 AASPSEKGAHPSGGADDVAKKPPPAPQQPPPPPAPHPQQHPQQHPQNQAHGKGGHRGGGG 74
Query: 259 GGWQHDSRDS 230
GG + S S
Sbjct: 75 GGGKSSSSSS 84
>pir||S33377 P63 protein - human
Length = 601
Score = 41.2 bits (95), Expect = 0.007
Identities = 27/70 (38%), Positives = 34/70 (48%), Gaps = 6/70 (8%)
Frame = -2
Query: 421 AGAPFEQVADPRDAKGVLASQPMHLP--PPPPPLPH-MSHLKQ---NYYSGEPGHDGQGY 260
A +P E+ A P +A +P P PPPPP PH H +Q N G+ GH G G
Sbjct: 15 AASPSEKGAHPSGGADDVAKKPPPAPQQPPPPPAPHPQQHPQQHPQNQAHGKGGHRGGGG 74
Query: 259 GGWQHDSRDS 230
GG + S S
Sbjct: 75 GGGKSSSSSS 84
>emb|CAA65506.1| cuticular collagen [Teladorsagia circumcincta]
Length = 284
Score = 39.7 bits (91), Expect = 0.020
Identities = 34/90 (37%), Positives = 37/90 (40%)
Frame = -2
Query: 481 GGDQSQLPPGFQGNYAQPWHAGAPFEQVADPRDAKGVLASQPMHLPPPPPPLPHMSHLKQ 302
GG PPG GN QP +AG+P A G L P PP PP P
Sbjct: 173 GGPGPIGPPGPPGNAGQPGNAGSP--------GAPGTLTQGPGSQGPPGPPGP------- 217
Query: 301 NYYSGEPGHDGQGYGGWQHDSRDSFGSPRL 212
G PG DGQ GG + RD G P L
Sbjct: 218 ---QGPPGPDGQP-GGPGNPGRD--GGPGL 241
>emb|CAA65507.1| cuticular collagen [Teladorsagia circumcincta]
Length = 284
Score = 37.7 bits (86), Expect = 0.076
Identities = 33/90 (36%), Positives = 36/90 (39%)
Frame = -2
Query: 481 GGDQSQLPPGFQGNYAQPWHAGAPFEQVADPRDAKGVLASQPMHLPPPPPPLPHMSHLKQ 302
GG PPG GN QP + G+P A G L P PP PP P
Sbjct: 173 GGPGPIGPPGPPGNAGQPGNPGSP--------GAPGTLTQGPGSQGPPGPPGP------- 217
Query: 301 NYYSGEPGHDGQGYGGWQHDSRDSFGSPRL 212
G PG DGQ GG + RD G P L
Sbjct: 218 ---QGPPGPDGQP-GGPGNPGRD--GGPGL 241
>ref|NP_037279.1| chloride channel 1; Chloride channel 1 (skeletal muscle) [Rattus
norvegicus] gi|544025|sp|P35524|CLC1_RAT Chloride
channel protein, skeletal muscle (Chloride channel
protein 1) (ClC-1) gi|103752|pir||S19595 chloride
channel protein - rat gi|57745|emb|CAA44683.1| skeletal
muscle chloride channel [Rattus norvegicus]
gi|228296|prf||1802386A Cl channel
Length = 994
Score = 37.4 bits (85), Expect = 0.099
Identities = 24/77 (31%), Positives = 30/77 (38%), Gaps = 7/77 (9%)
Frame = -2
Query: 505 RQDSTSDIGGDQSQLPPGFQGNYAQPWHAGAPFEQVA-------DPRDAKGVLASQPMHL 347
R + D+ S+LP Q A WH G E A + K L P
Sbjct: 670 RLKAAQDMARKLSELPYNGQAQLAGEWHPGGRPESFAFVDEDEDEDVSRKTELPQTPTPP 729
Query: 346 PPPPPPLPHMSHLKQNY 296
PPPPPPLP + +Y
Sbjct: 730 PPPPPPLPPQFPIAPSY 746
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,524,896
Number of Sequences: 1393205
Number of extensions: 11674640
Number of successful extensions: 81812
Number of sequences better than 10.0: 274
Number of HSP's better than 10.0 without gapping: 51403
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 72081
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 15652649358
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)