Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002507A_C01 KMC002507A_c01
(667 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
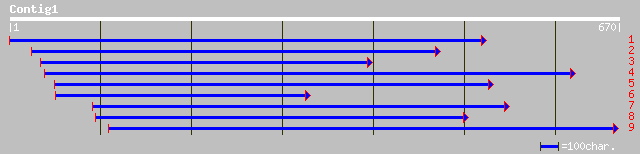
Sequences producing significant alignments: (bits) Value
emb|CAD40988.1| OSJNBa0072F16.12 [Oryza sativa (japonica cultiva... 44 5e-06
dbj|BAC53811.1| OsNAC2 protein [Oryza sativa] 44 5e-06
emb|CAA63101.1| NAM [Petunia x hybrida] gi|6066595|emb|CAA63102.... 43 7e-06
gb|AAK27685.1|AF347698_1 ADP-glucose pyrophosphorylase large sub... 50 4e-05
gb|AAM50521.1|AF510215_1 nam-like protein 18 [Petunia x hybrida] 40 4e-05
>emb|CAD40988.1| OSJNBa0072F16.12 [Oryza sativa (japonica cultivar-group)]
Length = 341
Score = 43.5 bits (101), Expect(2) = 5e-06
Identities = 17/28 (60%), Positives = 24/28 (85%)
Frame = -2
Query: 414 YYLPQKVLDSYFYAIAIADADLHKCEPW 331
+YL +KV D+ F A+A+A+ADL+KCEPW
Sbjct: 28 HYLAKKVADARFAALAVAEADLNKCEPW 55
Score = 28.5 bits (62), Expect(2) = 5e-06
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = -3
Query: 452 GFRFHPTNEELI 417
GFRFHPT+EELI
Sbjct: 15 GFRFHPTDEELI 26
>dbj|BAC53811.1| OsNAC2 protein [Oryza sativa]
Length = 334
Score = 43.5 bits (101), Expect(2) = 5e-06
Identities = 17/28 (60%), Positives = 24/28 (85%)
Frame = -2
Query: 414 YYLPQKVLDSYFYAIAIADADLHKCEPW 331
+YL +KV D+ F A+A+A+ADL+KCEPW
Sbjct: 19 HYLAKKVADARFAALAVAEADLNKCEPW 46
Score = 28.5 bits (62), Expect(2) = 5e-06
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = -3
Query: 452 GFRFHPTNEELI 417
GFRFHPT+EELI
Sbjct: 6 GFRFHPTDEELI 17
>emb|CAA63101.1| NAM [Petunia x hybrida] gi|6066595|emb|CAA63102.2| NAM [Petunia x
hybrida]
Length = 327
Score = 43.1 bits (100), Expect(2) = 7e-06
Identities = 19/28 (67%), Positives = 22/28 (77%)
Frame = -2
Query: 414 YYLPQKVLDSYFYAIAIADADLHKCEPW 331
YYL +KVLDS F AIA+ DL+KCEPW
Sbjct: 30 YYLLKKVLDSNFTGRAIAEVDLNKCEPW 57
Score = 28.5 bits (62), Expect(2) = 7e-06
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = -3
Query: 452 GFRFHPTNEELI 417
GFRFHPT+EELI
Sbjct: 17 GFRFHPTDEELI 28
>gb|AAK27685.1|AF347698_1 ADP-glucose pyrophosphorylase large subunit [Brassica rapa subsp.
pekinensis]
Length = 570
Score = 49.7 bits (117), Expect = 4e-05
Identities = 25/29 (86%), Positives = 27/29 (92%)
Frame = -2
Query: 561 VDTTVLGLSQEEAENKPSYIASMGVYIFK 475
VDTTVLGLS+EEAE KP YIASMGVY+FK
Sbjct: 301 VDTTVLGLSKEEAEKKP-YIASMGVYVFK 328
>gb|AAM50521.1|AF510215_1 nam-like protein 18 [Petunia x hybrida]
Length = 331
Score = 40.4 bits (93), Expect(2) = 4e-05
Identities = 16/28 (57%), Positives = 22/28 (78%)
Frame = -2
Query: 414 YYLPQKVLDSYFYAIAIADADLHKCEPW 331
+YL +KV D+ F+AIAI D D++K EPW
Sbjct: 34 HYLSKKVADNNFFAIAIGDVDMNKVEPW 61
Score = 28.5 bits (62), Expect(2) = 4e-05
Identities = 11/12 (91%), Positives = 12/12 (99%)
Frame = -3
Query: 452 GFRFHPTNEELI 417
GFRFHPT+EELI
Sbjct: 21 GFRFHPTDEELI 32
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 534,102,154
Number of Sequences: 1393205
Number of extensions: 11021677
Number of successful extensions: 37561
Number of sequences better than 10.0: 242
Number of HSP's better than 10.0 without gapping: 32801
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 36181
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 28855580904
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)