Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
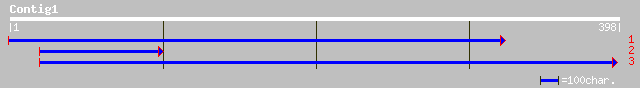
Query= KMC002504A_C01 KMC002504A_c01
(398 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T06371 probable UDP-glucuronosyltransferase (EC 2.4.1.-) - ... 106 8e-23
dbj|BAB86928.1| glucosyltransferase-10 [Vigna angularis] 95 2e-19
pir||D86356 hypothetical protein T16E15.4 - Arabidopsis thaliana... 89 1e-17
pir||F86356 T16E15.2 protein - Arabidopsis thaliana gi|9392678|g... 86 9e-17
ref|NP_564170.1| UDP-glucose glucosyltransferase, putative; prot... 86 9e-17
>pir||T06371 probable UDP-glucuronosyltransferase (EC 2.4.1.-) - garden pea
gi|2827992|gb|AAB99950.1| UDP-glucuronosyltransferase
[Pisum sativum]
Length = 347
Score = 106 bits (264), Expect = 8e-23
Identities = 46/57 (80%), Positives = 54/57 (94%)
Frame = -2
Query: 397 FFAEQQTNCRYCCEEWGIGLEIEDAKRDRVESLVRELMDGEKGKLMKENALKWKKLA 227
FFAEQQTNCR+CC EWGIGLEIEDAKRD++ESLV+E+++GEKGK MKE AL+WKKLA
Sbjct: 267 FFAEQQTNCRFCCHEWGIGLEIEDAKRDKIESLVKEMVEGEKGKEMKEKALEWKKLA 323
>dbj|BAB86928.1| glucosyltransferase-10 [Vigna angularis]
Length = 485
Score = 94.7 bits (234), Expect = 2e-19
Identities = 41/57 (71%), Positives = 48/57 (83%)
Frame = -2
Query: 397 FFAEQQTNCRYCCEEWGIGLEIEDAKRDRVESLVRELMDGEKGKLMKENALKWKKLA 227
FFAEQQTNCR+CC+EWGIG+EIED +RD +E LVR +MDGEKGK MK A+ WK LA
Sbjct: 397 FFAEQQTNCRFCCKEWGIGVEIEDVERDHIERLVRAMMDGEKGKDMKRKAVNWKILA 453
>pir||D86356 hypothetical protein T16E15.4 - Arabidopsis thaliana
gi|9392681|gb|AAF87258.1|AC068562_5 Strong similarity to
UDP-glucose glucosyltransferase from Arabidopsis
thaliana gb|AB016819 and contains a UDP-glucosyl
transferase PF|00201 domain
Length = 450
Score = 89.4 bits (220), Expect = 1e-17
Identities = 38/59 (64%), Positives = 52/59 (87%), Gaps = 1/59 (1%)
Frame = -2
Query: 397 FFAEQQTNCRYCCEEWGIGLEI-EDAKRDRVESLVRELMDGEKGKLMKENALKWKKLAH 224
FFAEQQTNC++CC+EWG+G+EI D KR+ VE++VRELMD EKGK M+E A++W++LA+
Sbjct: 367 FFAEQQTNCKFCCDEWGVGVEIGGDVKREEVETVVRELMDREKGKKMREKAVEWRRLAN 425
>pir||F86356 T16E15.2 protein - Arabidopsis thaliana
gi|9392678|gb|AAF87255.1|AC068562_2 Strong similarity to
UDP-glucose glucosyltransferase from Arabidopsis
thaliana gb|AB016819 and contains a UDP-glucosyl
transferase PF|00201 domain. ESTs gb|U74128,
gb|AA713257 come from this gene
Length = 479
Score = 86.3 bits (212), Expect = 9e-17
Identities = 37/58 (63%), Positives = 49/58 (83%), Gaps = 1/58 (1%)
Frame = -2
Query: 397 FFAEQQTNCRYCCEEWGIGLEI-EDAKRDRVESLVRELMDGEKGKLMKENALKWKKLA 227
FFAEQQTNC+YCC+EW +G+EI D +R+ VE LVRELMDG+KGK M++ A +W++LA
Sbjct: 396 FFAEQQTNCKYCCDEWEVGMEIGGDVRREEVEELVRELMDGDKGKKMRQKAEEWQRLA 453
>ref|NP_564170.1| UDP-glucose glucosyltransferase, putative; protein id: At1g22370.1,
supported by cDNA: gi_14532545, supported by cDNA:
gi_18655386 [Arabidopsis thaliana]
gi|14532546|gb|AAK64001.1| At1g22370/T16E15_3
[Arabidopsis thaliana] gi|18655387|gb|AAL76149.1|
At1g22370/T16E15_3 [Arabidopsis thaliana]
Length = 309
Score = 86.3 bits (212), Expect = 9e-17
Identities = 37/58 (63%), Positives = 49/58 (83%), Gaps = 1/58 (1%)
Frame = -2
Query: 397 FFAEQQTNCRYCCEEWGIGLEI-EDAKRDRVESLVRELMDGEKGKLMKENALKWKKLA 227
FFAEQQTNC+YCC+EW +G+EI D +R+ VE LVRELMDG+KGK M++ A +W++LA
Sbjct: 226 FFAEQQTNCKYCCDEWEVGMEIGGDVRREEVEELVRELMDGDKGKKMRQKAEEWQRLA 283
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 377,533,439
Number of Sequences: 1393205
Number of extensions: 8292504
Number of successful extensions: 24340
Number of sequences better than 10.0: 168
Number of HSP's better than 10.0 without gapping: 23427
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 24273
length of database: 448,689,247
effective HSP length: 108
effective length of database: 298,223,107
effective search space used: 7157354568
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)