Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002502A_C01 KMC002502A_c01
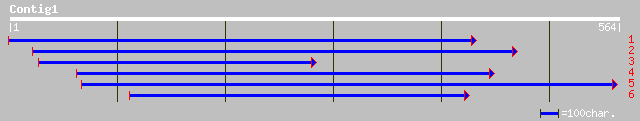
(557 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T45680 hypothetical protein F14P22.190 - Arabidopsis thalia... 45 6e-04
ref|NP_567071.1| putative protein; protein id: At3g58600.1, supp... 45 6e-04
gb|AAM63770.1| unknown [Arabidopsis thaliana] 45 6e-04
dbj|BAB17104.1| P0410E01.25 [Oryza sativa (japonica cultivar-gro... 33 1.8
sp|P45974|UBP5_HUMAN Ubiquitin carboxyl-terminal hydrolase 5 (Ub... 33 1.8
>pir||T45680 hypothetical protein F14P22.190 - Arabidopsis thaliana
gi|6735377|emb|CAB68198.1| putative protein [Arabidopsis
thaliana]
Length = 292
Score = 45.1 bits (105), Expect = 6e-04
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 4/63 (6%)
Frame = -1
Query: 554 VAAVQNSPTDSPTEFSPEKTSKLETSKTIQEDAEGEKPPENQS----TQDVVDDDFGDFQ 387
V Q SP+ P S +++S+ + T +E+ E EK + ++ +D DDDFGDFQ
Sbjct: 230 VTTAQKSPSSLPPSLSLQRSSEQQDLDTKREEEEEEKKEDLKAKDGGVEDAPDDDFGDFQ 289
Query: 386 AAG 378
AAG
Sbjct: 290 AAG 292
>ref|NP_567071.1| putative protein; protein id: At3g58600.1, supported by cDNA: 27.
[Arabidopsis thaliana] gi|25054905|gb|AAN71933.1|
unknown protein [Arabidopsis thaliana]
Length = 302
Score = 45.1 bits (105), Expect = 6e-04
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 4/63 (6%)
Frame = -1
Query: 554 VAAVQNSPTDSPTEFSPEKTSKLETSKTIQEDAEGEKPPENQS----TQDVVDDDFGDFQ 387
V Q SP+ P S +++S+ + T +E+ E EK + ++ +D DDDFGDFQ
Sbjct: 240 VTTAQKSPSSLPPSLSLQRSSEQQDLDTKREEEEEEKKEDLKAKDGGVEDAPDDDFGDFQ 299
Query: 386 AAG 378
AAG
Sbjct: 300 AAG 302
>gb|AAM63770.1| unknown [Arabidopsis thaliana]
Length = 293
Score = 45.1 bits (105), Expect = 6e-04
Identities = 24/63 (38%), Positives = 36/63 (57%), Gaps = 4/63 (6%)
Frame = -1
Query: 554 VAAVQNSPTDSPTEFSPEKTSKLETSKTIQEDAEGEKPPENQS----TQDVVDDDFGDFQ 387
V Q SP+ P S +++S+ + T +E+ E EK + ++ +D DDDFGDFQ
Sbjct: 231 VTTAQKSPSSLPPSLSLQRSSEQQDLDTKREEEEEEKKEDLKAKDGGVEDAPDDDFGDFQ 290
Query: 386 AAG 378
AAG
Sbjct: 291 AAG 293
>dbj|BAB17104.1| P0410E01.25 [Oryza sativa (japonica cultivar-group)]
Length = 245
Score = 33.5 bits (75), Expect = 1.8
Identities = 17/32 (53%), Positives = 20/32 (62%), Gaps = 6/32 (18%)
Frame = -1
Query: 455 EGEKPPENQST------QDVVDDDFGDFQAAG 378
+ E P+ Q T Q+ VDDDFGDFQAAG
Sbjct: 214 KAEAAPQEQPTAAEKAPQESVDDDFGDFQAAG 245
>sp|P45974|UBP5_HUMAN Ubiquitin carboxyl-terminal hydrolase 5 (Ubiquitin thiolesterase 5)
(Ubiquitin-specific processing protease 5)
(Deubiquitinating enzyme 5) (Isopeptidase T)
gi|1208744|gb|AAC50465.1| isopeptidase T
gi|1732412|gb|AAB51315.1| isopeptidase T [Homo sapiens]
gi|13477329|gb|AAH05139.1|AAH05139 Similar to ubiquitin
specific protease 5 (isopeptidase T) [Homo sapiens]
Length = 858
Score = 33.5 bits (75), Expect = 1.8
Identities = 19/49 (38%), Positives = 27/49 (54%), Gaps = 7/49 (14%)
Frame = -1
Query: 542 QNSPTDSPTEFSPEKTSKL-------ETSKTIQEDAEGEKPPENQSTQD 417
QN+PTD +FS + +KL E SK + E +GE+ PE + QD
Sbjct: 363 QNAPTDPTQDFSTQ-VAKLGHGLLSGEYSKPVPESGDGERVPEQKEVQD 410
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 433,475,265
Number of Sequences: 1393205
Number of extensions: 8425079
Number of successful extensions: 31143
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 30267
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31102
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)