Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC002501A_C01 KMC002501A_c01
(672 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
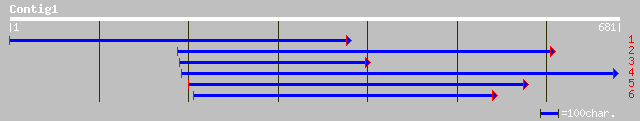
ref|NP_176319.1| hypothetical protein; protein id: At1g61240.1 [... 128 6e-29
gb|AAN63500.1|AF317555_1 lysine ketoglutarate reductase trans-sp... 125 7e-28
ref|NP_172583.2| hypothetical protein; protein id: At1g11170.1, ... 125 7e-28
pir||F86245 hypothetical protein [imported] - Arabidopsis thalia... 121 1e-26
dbj|BAC22542.1| hypothetical protein~similar to Arabidopsis thal... 113 2e-24
>ref|NP_176319.1| hypothetical protein; protein id: At1g61240.1 [Arabidopsis
thaliana] gi|26449863|dbj|BAC42054.1| unknown protein
[Arabidopsis thaliana]
Length = 425
Score = 128 bits (322), Expect = 6e-29
Identities = 62/113 (54%), Positives = 78/113 (68%), Gaps = 8/113 (7%)
Frame = -1
Query: 672 GWGVDIKLGYCAQGDRTQNVGIVDSEYVVHNAIQTLGGSSHDHKAKKHR--------ASA 517
GWG+D+KLGYCAQGDR++ VGIVDSEY+ H IQTLGGS + K R ++
Sbjct: 298 GWGMDMKLGYCAQGDRSKKVGIVDSEYIFHQGIQTLGGSGYPDKKNSARSGVNRRRGSAT 357
Query: 516 LDARTEIRRQSSWELQIFKERWNKAVAEDRNWVDPFKRDQRRLLQRRNRQRFS 358
D+RTEIRRQS+WELQ FKERWN+AVAED+ WV+ + R+ R +R S
Sbjct: 358 FDSRTEIRRQSTWELQAFKERWNQAVAEDKYWVERSSPSRNRIGNNRRLKRSS 410
>gb|AAN63500.1|AF317555_1 lysine ketoglutarate reductase trans-splicing related 1
[Arabidopsis thaliana]
Length = 358
Score = 125 bits (313), Expect = 7e-28
Identities = 61/113 (53%), Positives = 77/113 (67%), Gaps = 8/113 (7%)
Frame = -1
Query: 672 GWGVDIKLGYCAQGDRTQNVGIVDSEYVVHNAIQTLGGSSHDHKAKKHR--------ASA 517
GWG+D+KLG CAQGDR++ VGIVDSEY+ H IQTLGGS + K R ++
Sbjct: 231 GWGMDMKLGSCAQGDRSKKVGIVDSEYIFHQGIQTLGGSGYPDKKNSARSGVNRRRGSAT 290
Query: 516 LDARTEIRRQSSWELQIFKERWNKAVAEDRNWVDPFKRDQRRLLQRRNRQRFS 358
D+RTEIRRQS+WELQ FKERWN+AVAED+ WV+ + R+ R +R S
Sbjct: 291 FDSRTEIRRQSTWELQAFKERWNQAVAEDKYWVERSSPSRNRIGNNRRLKRSS 343
>ref|NP_172583.2| hypothetical protein; protein id: At1g11170.1, supported by cDNA:
gi_18700104 [Arabidopsis thaliana]
gi|18700105|gb|AAL77664.1| At1g11170/T28P6_16
[Arabidopsis thaliana] gi|21700791|gb|AAM70519.1|
At1g11170/T28P6_16 [Arabidopsis thaliana]
Length = 438
Score = 125 bits (313), Expect = 7e-28
Identities = 58/94 (61%), Positives = 71/94 (74%), Gaps = 7/94 (7%)
Frame = -1
Query: 672 GWGVDIKLGYCAQGDRTQNVGIVDSEYVVHNAIQTLGGS-------SHDHKAKKHRASAL 514
GWG+D+KLGYCAQGDRT+NVGIVDSEY++H IQTLG S + D ++ +
Sbjct: 301 GWGMDMKLGYCAQGDRTKNVGIVDSEYILHQGIQTLGESVPEKKKTARDVGTRRQGHTTF 360
Query: 513 DARTEIRRQSSWELQIFKERWNKAVAEDRNWVDP 412
D+RTEIRRQS+WELQ FKERW+KAV ED W+DP
Sbjct: 361 DSRTEIRRQSTWELQTFKERWSKAVEEDIKWIDP 394
>pir||F86245 hypothetical protein [imported] - Arabidopsis thaliana
gi|5734733|gb|AAD49998.1|AC007259_11 Hypothetical
protein [Arabidopsis thaliana]
Length = 266
Score = 121 bits (303), Expect = 1e-26
Identities = 57/87 (65%), Positives = 67/87 (76%)
Frame = -1
Query: 672 GWGVDIKLGYCAQGDRTQNVGIVDSEYVVHNAIQTLGGSSHDHKAKKHRASALDARTEIR 493
GWG+D+KLGYCAQGDRT+NVGIVDSEY++H IQTLG S + K +A D T IR
Sbjct: 141 GWGMDMKLGYCAQGDRTKNVGIVDSEYILHQGIQTLGESVPEKK-----KTARDVGTRIR 195
Query: 492 RQSSWELQIFKERWNKAVAEDRNWVDP 412
RQS+WELQ FKERW+KAV ED W+DP
Sbjct: 196 RQSTWELQTFKERWSKAVEEDIKWIDP 222
>dbj|BAC22542.1| hypothetical protein~similar to Arabidopsis thaliana chromosome 1,
At1g11170 [Oryza sativa (japonica cultivar-group)]
Length = 300
Score = 113 bits (283), Expect = 2e-24
Identities = 58/118 (49%), Positives = 75/118 (63%), Gaps = 16/118 (13%)
Frame = -1
Query: 672 GWGVDIKLGYCAQGDRTQNVGIVDSEYVVHNAIQTLGGSS------------HDHKAKKH 529
GWG+D K GYCAQGDRT+N+G+VDSEY+VH +QTLGG S H A+
Sbjct: 184 GWGIDYKFGYCAQGDRTKNIGVVDSEYIVHRGVQTLGGPSVKRSHGKNNDPLHQKTAEAQ 243
Query: 528 R----ASALDARTEIRRQSSWELQIFKERWNKAVAEDRNWVDPFKRDQRRLLQRRNRQ 367
+ + LD RT++RR S EL+ F++RW +A EDR WVDPF R RR +R +RQ
Sbjct: 244 QQMRVKAGLDMRTKVRRYSRSELRDFQKRWERATREDRAWVDPFAR-PRRKRKRTDRQ 300
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 534,116,351
Number of Sequences: 1393205
Number of extensions: 11041893
Number of successful extensions: 27894
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 26821
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27852
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29421376608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)