Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
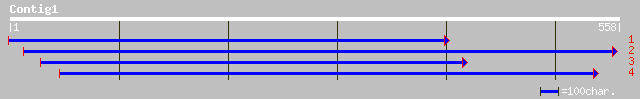
Query= KMC002492A_C01 KMC002492A_c01
(532 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAB63264.3| calcium-binding protein [Lotus japonicus] 121 1e-28
ref|NP_187405.1| putative calmodulin; protein id: At3g07490.1 [A... 108 1e-23
ref|NP_193022.1| putative calmodulin; protein id: At4g12860.1 [A... 106 5e-23
ref|NP_172089.1| calcium-binding protein, putative; protein id: ... 103 7e-22
ref|NP_565996.1| putative calcium binding protein; protein id: A... 98 4e-20
>emb|CAB63264.3| calcium-binding protein [Lotus japonicus]
Length = 230
Score = 121 bits (304), Expect(2) = 1e-28
Identities = 59/59 (100%), Positives = 59/59 (100%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFK 354
REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFK
Sbjct: 160 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFK 218
Score = 48.1 bits (113), Expect = 6e-05
Identities = 26/58 (44%), Positives = 37/58 (62%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEF 357
+ F +FD+NGDG IT +EL L +LGI ++ +MI ++DV+GDG VD EF
Sbjct: 87 KRVFQMFDRNGDGRITKKELNDSLENLGI--FIPDKELTQMIERIDVNGDGCVDIDEF 142
Score = 26.2 bits (56), Expect(2) = 1e-28
Identities = 11/11 (100%), Positives = 11/11 (100%)
Frame = -1
Query: 349 MMKGGGFSALT 317
MMKGGGFSALT
Sbjct: 220 MMKGGGFSALT 230
>ref|NP_187405.1| putative calmodulin; protein id: At3g07490.1 [Arabidopsis thaliana]
gi|6041859|gb|AAF02168.1|AC009853_28 putative calmodulin
[Arabidopsis thaliana]
Length = 153
Score = 108 bits (269), Expect(2) = 1e-23
Identities = 51/59 (86%), Positives = 57/59 (96%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFK 354
REAFNVFDQN DGFITVEELR+VLASLG+KQGRT+EDCK+MI KVDVDGDGMV++KEFK
Sbjct: 80 REAFNVFDQNRDGFITVEELRSVLASLGLKQGRTLEDCKRMISKVDVDGDGMVNFKEFK 138
Score = 50.1 bits (118), Expect = 2e-05
Identities = 27/55 (49%), Positives = 37/55 (67%)
Frame = -3
Query: 521 FNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEF 357
F +FD+NGDG IT +EL L +LGI +D +MI K+D++GDG VD +EF
Sbjct: 10 FQMFDRNGDGKITKQELNDSLENLGIY--IPDKDLVQMIEKIDLNGDGYVDIEEF 62
Score = 23.1 bits (48), Expect(2) = 1e-23
Identities = 9/10 (90%), Positives = 10/10 (100%)
Frame = -1
Query: 349 MMKGGGFSAL 320
MMKGGGF+AL
Sbjct: 140 MMKGGGFAAL 149
>ref|NP_193022.1| putative calmodulin; protein id: At4g12860.1 [Arabidopsis thaliana]
gi|7441488|pir||T06644 calmodulin homolog T20K18.210 -
Arabidopsis thaliana gi|4586262|emb|CAB41003.1| putative
calmodulin [Arabidopsis thaliana]
gi|7267988|emb|CAB78328.1| putative calmodulin
[Arabidopsis thaliana]
Length = 152
Score = 106 bits (265), Expect(2) = 5e-23
Identities = 50/59 (84%), Positives = 56/59 (94%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFK 354
REAF VFDQNGDGFIT EELR+VLAS+G+KQGRT+EDCKKMI KVDVDGDGMV++KEFK
Sbjct: 80 REAFRVFDQNGDGFITDEELRSVLASMGLKQGRTLEDCKKMISKVDVDGDGMVNFKEFK 138
Score = 46.2 bits (108), Expect = 2e-04
Identities = 24/55 (43%), Positives = 33/55 (59%)
Frame = -3
Query: 521 FNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEF 357
F +FD+NGDG I EL+ S+GI + +MI K+DV+GDG +D EF
Sbjct: 10 FQMFDKNGDGKIAKNELKDFFKSVGIMVPE--NEINEMIAKMDVNGDGAMDIDEF 62
Score = 22.3 bits (46), Expect(2) = 5e-23
Identities = 8/11 (72%), Positives = 11/11 (99%)
Frame = -1
Query: 349 MMKGGGFSALT 317
MM+GGGF+AL+
Sbjct: 140 MMRGGGFAALS 150
>ref|NP_172089.1| calcium-binding protein, putative; protein id: At1g05990.1,
supported by cDNA: gi_12083339 [Arabidopsis thaliana]
gi|25295760|pir||H86194 hypothetical protein [imported]
- Arabidopsis thaliana
gi|8810461|gb|AAF80122.1|AC024174_4 Contains similarity
to a calcium-binding protein from Lotus japonicus
gi|6580549 and contains a EF hand PF|00036 domain. EST
gb|T46471 comes from this gene. [Arabidopsis thaliana]
gi|12083340|gb|AAG48829.1|AF332466_1 putative
calcium-binding protein [Arabidopsis thaliana]
Length = 150
Score = 103 bits (256), Expect(2) = 7e-22
Identities = 46/59 (77%), Positives = 56/59 (93%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFK 354
+EAFNVFDQNGDGFITV+EL+ VL+SLG+KQG+T++DCKKMI KVDVDGDG V+YKEF+
Sbjct: 81 KEAFNVFDQNGDGFITVDELKAVLSSLGLKQGKTLDDCKKMIKKVDVDGDGRVNYKEFR 139
Score = 51.2 bits (121), Expect = 7e-06
Identities = 28/58 (48%), Positives = 37/58 (63%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEF 357
+ F +FD+NGDG IT +EL L SLGI ++ +MI K+DV+GDG VD EF
Sbjct: 7 KRVFQMFDKNGDGTITGKELSETLRSLGIY--IPDKELTQMIEKIDVNGDGCVDIDEF 62
Score = 21.9 bits (45), Expect(2) = 7e-22
Identities = 8/10 (80%), Positives = 10/10 (100%)
Frame = -1
Query: 349 MMKGGGFSAL 320
MMKGGGF++L
Sbjct: 141 MMKGGGFNSL 150
>ref|NP_565996.1| putative calcium binding protein; protein id: At2g43290.1,
supported by cDNA: 31535., supported by cDNA:
gi_17065477, supported by cDNA: gi_20148490, supported
by cDNA: gi_9965746 [Arabidopsis thaliana]
gi|9965747|gb|AAG10150.1|AF250344_1 calmodulin-like MSS3
[Arabidopsis thaliana] gi|17065478|gb|AAL32893.1|
putative Ca2+-binding protein [Arabidopsis thaliana]
gi|20148491|gb|AAM10136.1| putative Ca2+-binding protein
[Arabidopsis thaliana] gi|20196862|gb|AAB64310.2|
putative calcium binding protein [Arabidopsis thaliana]
gi|20197147|gb|AAM14938.1| putative calcium binding
protein [Arabidopsis thaliana]
gi|21592699|gb|AAM64648.1| putative calcium binding
protein [Arabidopsis thaliana]
Length = 215
Score = 97.8 bits (242), Expect(2) = 4e-20
Identities = 43/58 (74%), Positives = 55/58 (94%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEF 357
++AFNVFDQ+GDGFITVEEL++V+ASLG+KQG+T++ CKKMIM+VD DGDG V+YKEF
Sbjct: 145 KDAFNVFDQDGDGFITVEELKSVMASLGLKQGKTLDGCKKMIMQVDADGDGRVNYKEF 202
Score = 51.6 bits (122), Expect = 6e-06
Identities = 28/59 (47%), Positives = 37/59 (62%)
Frame = -3
Query: 530 REAFNVFDQNGDGFITVEELRTVLASLGIKQGRTVEDCKKMIMKVDVDGDGMVDYKEFK 354
+ F +FD+NGDG IT EEL L +LGI +D +MI K+D +GDG VD EF+
Sbjct: 67 KRVFQMFDKNGDGRITKEELNDSLENLGIY--IPDKDLTQMIHKIDANGDGCVDIDEFE 123
Score = 21.6 bits (44), Expect(2) = 4e-20
Identities = 8/9 (88%), Positives = 9/9 (99%)
Frame = -1
Query: 349 MMKGGGFSA 323
MMKGGGFS+
Sbjct: 205 MMKGGGFSS 213
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 544,968,173
Number of Sequences: 1393205
Number of extensions: 13261796
Number of successful extensions: 35847
Number of sequences better than 10.0: 1570
Number of HSP's better than 10.0 without gapping: 31804
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34631
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17596710992
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)